FOS Rescues Neuronal Differentiation of Sox2-Deleted Neural Stem Cells by Genome-Wide Regulation of Common SOX2 and AP1(FOS-JUN) Target Genes
Abstract
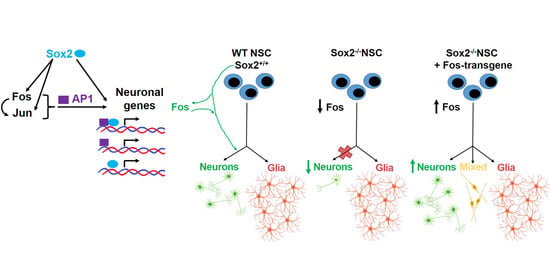
:1. Introduction
2. Materials and Methods
2.1. Primary Ex-Vivo Neural Stem/Progenitor Cell Cultures
2.2. RNA-seq
2.3. Lentiviral Constructs
2.4. Sox2-del Transduction with Lentiviral Constructs Encoding Sox2, Socs3 and Fos
2.5. RT-PCR Analysis of Sox2, Fos, and Socs3
2.6. NSC Differentiation
2.7. Immunocytochemistry
2.8. GO Enrichment Analysis
2.9. CUT&RUN
3. Results
3.1. Lentiviral Expression of Sox2 in Sox2-Deleted NSC Rescues Their Ability to Generate Neurons upon Induction of Differentiation
3.2. Lentiviral Expression of Fos in Sox2-del NSC Rescues Their Ability to Generate Neurons upon Induction of Differentiation
3.3. Lentiviral Expression of Socs3 in Sox2-del NSC Inhibits the Genesis of Glial Cells upon Induction of Differentiation
3.4. RNA-seq Demonstrates That Genes Downregulated in Undifferentiated Sox2-del NSC Are Enriched in Genes Involved in Neuronal Differentiation
3.5. CUT&RUN Reveals Broad Genome-Wide Co-Occupancy of SOX2 and the AP1 Complex in NSC
3.6. The SOX2/AP1 Duet Regulates a Large Fraction of Genes Expressed in NSC
3.7. SOX2 and the AP1 Complex Regulate Genes Involved in Neuronal Differentiation
4. Discussion
4.1. Fos Overexpression in Sox2-del Cells Favours Initial Neuronal Differentiation of a Glial/Neuronal Progenitor
4.2. FOS Binds, in NSC, to a Large Proportion of Expressed Genes Close to SOX2-Bound Regions
4.3. SOX Transcription Factors Orchestrate Signalling Pathways by Promiscuous Interactions
4.4. A Network of Interactions Directing Neuronal versus Glial Differentiation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bertolini, J.; Mercurio, S.; Favaro, R.; Mariani, J.; Ottolenghi, S.; Nicolis, S.K. Sox2-dependent regulation of neural stem cells and CNS development. In Sox2, Biology and Role in Development and Disease; Kondoh, H., Lovell-Badge, R., Eds.; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Mercurio, S.; Serra, L.; Nicolis, S.K. More than just Stem Cells: Functional Roles of the Transcription Factor Sox2 in Differentiated Glia and Neurons. Int. J. Mol. Sci. 2019, 20, 4540. [Google Scholar] [CrossRef] [Green Version]
- Favaro, R.; Valotta, M.; Ferri, A.L.; Latorre, E.; Mariani, J.; Giachino, C.; Lancini, C.; Tosetti, V.; Ottolenghi, S.; Taylor, V.; et al. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat. Neurosci. 2009, 12, 1248–1256. [Google Scholar] [CrossRef]
- Ferri, A.; Favaro, R.; Beccari, L.; Bertolini, J.; Mercurio, S.; Nieto-Lopez, F.; Verzeroli, C.; La Regina, F.; De Pietri Tonelli, D.; Ottolenghi, S.; et al. Sox2 is required for embryonic development of the ventral telencephalon through the activation of the ventral determinants Nkx2.1 and Shh. Development 2013, 140, 1250–1261. [Google Scholar] [CrossRef] [Green Version]
- Ferri, A.L.; Cavallaro, M.; Braida, D.; Di Cristofano, A.; Canta, A.; Vezzani, A.; Ottolenghi, S.; Pandolfi, P.P.; Sala, M.; DeBiasi, S.; et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 2004, 131, 3805–3819. [Google Scholar] [CrossRef] [Green Version]
- Mercurio, S.; Alberti, C.; Serra, L.; Meneghini, S.; Berico, P.; Bertolini, J.; Becchetti, A.; Nicolis, S.K. An early Sox2-dependent gene expression programme required for hippocampal dentate gyrus development. Open Biol. 2021, in press. [Google Scholar] [CrossRef]
- Suh, H.; Consiglio, A.; Ray, J.; Sawai, T.; D’Amour, K.A.; Gage, F.H. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell 2007, 1, 515–528. [Google Scholar] [CrossRef] [Green Version]
- Bertolini, J.A.; Favaro, R.; Zhu, Y.; Pagin, M.; Ngan, C.Y.; Wong, C.H.; Tjong, H.; Vermunt, M.W.; Martynoga, B.; Barone, C.; et al. Mapping the Global Chromatin Connectivity Network for Sox2 Function in Neural Stem Cell Maintenance. Cell Stem Cell 2019, 24, 462–476.e466. [Google Scholar] [CrossRef] [Green Version]
- Pagin, M.; Pernebrink, M.; Giubbolini, S.; Barone, C.; Sambruni, G.; Zhu, Y.; Chiara, M.; Ottolenghi, S.; Pavesi, G.; Wei, C.L.; et al. Sox2 controls neural stem cell self-renewal through a Fos-centered gene regulatory network. Stem Cells 2021. [Google Scholar] [CrossRef]
- Cavallaro, M.; Mariani, J.; Lancini, C.; Latorre, E.; Caccia, R.; Gullo, F.; Valotta, M.; DeBiasi, S.; Spinardi, L.; Ronchi, A.; et al. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development 2008, 135, 541–557. [Google Scholar] [CrossRef] [Green Version]
- Mercurio, S.; Serra, L.; Motta, A.; Gesuita, L.; Sanchez-Arrones, L.; Inverardi, F.; Foglio, B.; Barone, C.; Kaimakis, P.; Martynoga, B.; et al. Sox2 Acts in Thalamic Neurons to Control the Development of Retina-Thalamus-Cortex Connectivity. iScience 2019, 15, 257–273. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wong, C.H.; Birnbaum, R.Y.; Li, G.; Favaro, R.; Ngan, C.Y.; Lim, J.; Tai, E.; Poh, H.M.; Wong, E.; et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 2013, 504, 306–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbarani, G.; Fugazza, C.; Barabino, S.M.L.; Ronchi, A.E. SOX6 blocks the proliferation of BCR-ABL1(+) and JAK2V617F(+) leukemic cells. Sci. Rep. 2019, 9, 3388. [Google Scholar] [CrossRef] [PubMed]
- Jagga, B.; Edwards, M.; Pagin, M.; Wagstaff, K.M.; Aragao, D.; Roman, N.; Nanson, J.D.; Raidal, S.R.; Dominado, N.; Stewart, M.; et al. Structural basis for nuclear import selectivity of pioneer transcription factor SOX2. Nat. Commun. 2021, 12, 28. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010, 32, 11.7.1–11.7.14. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meers, M.P.; Tenenbaum, D.; Henikoff, S. Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin 2019, 12, 42. [Google Scholar] [CrossRef] [Green Version]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [Green Version]
- McLean, C.Y.; Bristor, D.; Hiller, M.; Clarke, S.L.; Schaar, B.T.; Lowe, C.B.; Wenger, A.M.; Bejerano, G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010, 28, 495–501. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skene, P.J.; Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Farioli-Vecchioli, S.; Saraulli, D.; Costanzi, M.; Pacioni, S.; Cina, I.; Aceti, M.; Micheli, L.; Bacci, A.; Cestari, V.; Tirone, F. The timing of differentiation of adult hippocampal neurons is crucial for spatial memory. PLoS Biol. 2008, 6, e246. [Google Scholar] [CrossRef] [PubMed]
- Micheli, L.; Ceccarelli, M.; Farioli-Vecchioli, S.; Tirone, F. Control of the Normal and Pathological Development of Neural Stem and Progenitor Cells by the PC3/Tis21/Btg2 and Btg1 Genes. J. Cell Physiol. 2015, 230, 2881–2890. [Google Scholar] [CrossRef]
- Tirone, F.; Farioli-Vecchioli, S.; Micheli, L.; Ceccarelli, M.; Leonardi, L. Genetic control of adult neurogenesis: Interplay of differentiation, proliferation and survival modulates new neurons function, and memory circuits. Front. Cell Neurosci. 2013, 7, 59. [Google Scholar] [CrossRef] [Green Version]
- Karakatsani, A.; Marichal, N.; Urban, S.; Kalamakis, G.; Ghanem, A.; Schick, A.; Zhang, Y.; Conzelmann, K.K.; Ruegg, M.A.; Berninger, B.; et al. Neuronal LRP4 regulates synapse formation in the developing CNS. Development 2017, 144, 4604–4615. [Google Scholar] [CrossRef] [Green Version]
- Julca, D.M.; Diaz, J.; Berger, S.; Leon, E. MAP1B related syndrome: Case presentation and review of literature. Am. J. Med. Genet. A 2019, 179, 1703–1708. [Google Scholar] [CrossRef]
- Sole, L.; Wagnon, J.L.; Akin, E.J.; Meisler, M.H.; Tamkun, M.M. The MAP1B Binding Domain of Nav1.6 Is Required for Stable Expression at the Axon Initial Segment. J. Neurosci. 2019, 39, 4238–4251. [Google Scholar] [CrossRef] [Green Version]
- Walters, G.B.; Gustafsson, O.; Sveinbjornsson, G.; Eiriksdottir, V.K.; Agustsdottir, A.B.; Jonsdottir, G.A.; Steinberg, S.; Gunnarsson, A.F.; Magnusson, M.I.; Unnsteinsdottir, U.; et al. MAP1B mutations cause intellectual disability and extensive white matter deficit. Nat. Commun. 2018, 9, 3456. [Google Scholar] [CrossRef] [PubMed]
- Jiao, H.F.; Sun, X.D.; Bates, R.; Xiong, L.; Zhang, L.; Liu, F.; Li, L.; Zhang, H.S.; Wang, S.Q.; Xiong, M.T.; et al. Transmembrane protein 108 is required for glutamatergic transmission in dentate gyrus. Proc. Natl. Acad. Sci. USA 2017, 114, 1177–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Lin, D.; Zhong, Y.; Luo, B.; Liu, S.; Fei, E.; Lai, X.; Zou, S.; Wang, S. Transmembrane protein 108 involves in adult neurogenesis in the hippocampal dentate gyrus. Cell Biosci. 2019, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Briscoe, J.; Sussel, L.; Serup, P.; Hartigan-O’Connor, D.; Jessell, T.M.; Rubenstein, J.L.; Ericson, J. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature 1999, 398, 622–627. [Google Scholar] [CrossRef]
- Cheng, L.; Chen, C.L.; Luo, P.; Tan, M.; Qiu, M.; Johnson, R.; Ma, Q. Lmx1b, Pet-1, and Nkx2.2 coordinately specify serotonergic neurotransmitter phenotype. J. Neurosci. 2003, 23, 9961–9967. [Google Scholar] [CrossRef] [Green Version]
- Jarrar, W.; Dias, J.M.; Ericson, J.; Arnold, H.H.; Holz, A. Nkx2.2 and Nkx2.9 are the key regulators to determine cell fate of branchial and visceral motor neurons in caudal hindbrain. PLoS ONE 2015, 10, e0124408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, R.S.; Liu, P.P.; Xi, F.; Wang, W.H.; Tang, G.B.; Wang, R.Y.; Saijilafu; Liu, C.M. Wnt3 and Gata4 regulate axon regeneration in adult mouse DRG neurons. Biochem. Biophys. Res. Commun. 2018, 499, 246–252. [Google Scholar] [CrossRef]
- Chen, J.L.; Chang, C.H.; Tsai, J.W. Gli2 Rescues Delays in Brain Development Induced by Kif3a Dysfunction. Cereb. Cortex 2019, 29, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.S.; Miyoshi, G.; Hanashima, C. Sensory cortex wiring requires preselection of short- and long-range projection neurons through an Egr-Foxg1-COUP-TFI network. Nat. Commun. 2019, 10, 3581. [Google Scholar] [CrossRef] [Green Version]
- Hartwig, C.; Veske, A.; Krejcova, S.; Rosenberger, G.; Finckh, U. Plexin B3 promotes neurite outgrowth, interacts homophilically, and interacts with Rin. BMC Neurosci. 2005, 6, 53. [Google Scholar] [CrossRef] [Green Version]
- Buckley, D.M.; Burroughs-Garcia, J.; Kriks, S.; Lewandoski, M.; Waters, S.T. Gbx1 and Gbx2 Are Essential for Normal Patterning and Development of Interneurons and Motor Neurons in the Embryonic Spinal Cord. J. Dev. Biol. 2020, 8, 9. [Google Scholar] [CrossRef] [Green Version]
- Pei, Z.; Wang, B.; Chen, G.; Nagao, M.; Nakafuku, M.; Campbell, K. Homeobox genes Gsx1 and Gsx2 differentially regulate telencephalic progenitor maturation. Proc. Natl. Acad. Sci. USA 2011, 108, 1675–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Long, J.E.; Flandin, P.; Pla, R.; Waclaw, R.R.; Campbell, K.; Rubenstein, J.L. Loss of Gsx1 and Gsx2 function rescues distinct phenotypes in Dlx1/2 mutants. J. Comp. Neurol. 2013, 521, 1561–1584. [Google Scholar] [CrossRef] [Green Version]
- Meng, Q.; Xia, Y. c-Jun, at the crossroad of the signaling network. Protein Cell 2011, 2, 889–898. [Google Scholar] [CrossRef] [Green Version]
- Hor, H.; Francescatto, L.; Bartesaghi, L.; Ortega-Cubero, S.; Kousi, M.; Lorenzo-Betancor, O.; Jimenez-Jimenez, F.J.; Gironell, A.; Clarimon, J.; Drechsel, O.; et al. Missense mutations in TENM4, a regulator of axon guidance and central myelination, cause essential tremor. Hum. Mol. Genet. 2015, 24, 5677–5686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Hon, C.C.; Carninci, P. Expanded ENCODE delivers invaluable genomic encyclopedia. Nature 2020, 583, 685–686. [Google Scholar] [CrossRef]
- Partridge, E.C.; Chhetri, S.B.; Prokop, J.W.; Ramaker, R.C.; Jansen, C.S.; Goh, S.T.; Mackiewicz, M.; Newberry, K.M.; Brandsmeier, L.A.; Meadows, S.K.; et al. Occupancy maps of 208 chromatin-associated proteins in one human cell type. Nature 2020, 583, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Vierbuchen, T.; Ling, E.; Cowley, C.J.; Couch, C.H.; Wang, X.; Harmin, D.A.; Roberts, C.W.M.; Greenberg, M.E. AP-1 Transcription Factors and the BAF Complex Mediate Signal-Dependent Enhancer Selection. Mol. Cell 2017, 68, 1067–1082.e1012. [Google Scholar] [CrossRef] [Green Version]
- Pan, W.; Jin, Y.; Chen, J.; Rottier, R.J.; Steel, K.P.; Kiernan, A.E. Ectopic expression of activated notch or SOX2 reveals similar and unique roles in the development of the sensory cell progenitors in the mammalian inner ear. J. Neurosci. 2013, 33, 16146–16157. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Yang, X.; Sun, Y.; Yu, H.; Zhang, Y.; Jin, Y. Comprehensive profiling reveals mechanisms of SOX2-mediated cell fate specification in human ESCs and NPCs. Cell Res. 2016, 26, 171–189. [Google Scholar] [CrossRef]
- Heavner, W.E.; Andoniadou, C.L.; Pevny, L.H. Establishment of the neurogenic boundary of the mouse retina requires cooperation of SOX2 and WNT signaling. Neural. Dev. 2014, 9, 27. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Chaturvedi, P.; Rankin, S.A.; Fish, M.B.; Wlizla, M.; Paraiso, K.D.; MacDonald, M.; Chen, X.; Weirauch, M.T.; Blitz, I.L.; et al. Sox17 and beta-catenin co-occupy Wnt-responsive enhancers to govern the endoderm gene regulatory network. eLife 2020, 9. [Google Scholar] [CrossRef]
- Soderholm, S.; Cantu, C. The WNT/beta-catenin dependent transcription: A tissue-specific business. Wiley Interdiscip Rev. Syst. Biol. Med. 2020, e1511. [Google Scholar] [CrossRef]
- Auwerx, J.; Sassone-Corsi, P. AP-1 (Fos-Jun) regulation by IP-1: Effect of signal transduction pathways and cell growth. Oncogene 1992, 7, 2271–2280. [Google Scholar]
- Bergsland, M.; Ramsköld, D.; Zaouter, C.; Klum, S.; Sandberg, R.; Muhr, J. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011, 25, 2453–2464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corsinotti, A.; Wong, F.C.; Tatar, T.; Szczerbinska, I.; Halbritter, F.; Colby, D.; Gogolok, S.; Pantier, R.; Liggat, K.; Mirfazeli, E.S.; et al. Distinct SoxB1 networks are required for naive and primed pluripotency. eLife 2017, 6. [Google Scholar] [CrossRef] [Green Version]
- Barclay, J.L.; Anderson, S.T.; Waters, M.J.; Curlewis, J.D. Regulation of suppressor of cytokine signaling 3 (SOC3) by growth hormone in pro-B cells. Mol. Endocrinol. 2007, 21, 2503–2515. [Google Scholar] [CrossRef] [PubMed]
- Barclay, J.L.; Anderson, S.T.; Waters, M.J.; Curlewis, J.D. Characterization of the SOCS3 promoter response to prostaglandin E2 in T47D cells. Mol. Endocrinol. 2007, 21, 2516–2528. [Google Scholar] [CrossRef] [Green Version]
- Cao, F.; Hata, R.; Zhu, P.; Ma, Y.J.; Tanaka, J.; Hanakawa, Y.; Hashimoto, K.; Niinobe, M.; Yoshikawa, K.; Sakanaka, M. Overexpression of SOCS3 inhibits astrogliogenesis and promotes maintenance of neural stem cells. J. Neurochem. 2006, 98, 459–470. [Google Scholar] [CrossRef]
- Fukuda, S.; Abematsu, M.; Mori, H.; Yanagisawa, M.; Kagawa, T.; Nakashima, K.; Yoshimura, A.; Taga, T. Potentiation of astrogliogenesis by STAT3-mediated activation of bone morphogenetic protein-Smad signaling in neural stem cells. Mol. Cell Biol. 2007, 27, 4931–4937. [Google Scholar] [CrossRef] [Green Version]
- Velazquez, F.N.; Caputto, B.L.; Boussin, F.D. c-Fos importance for brain development. Aging (Albany N.Y.) 2015, 7, 1028–1029. [Google Scholar] [CrossRef] [Green Version]
- Velazquez, F.N.; Prucca, C.G.; Etienne, O.; D’Astolfo, D.S.; Silvestre, D.C.; Boussin, F.D.; Caputto, B.L. Brain development is impaired in c-fos -/- mice. Oncotarget 2015, 6, 16883–16901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emery, B.; Merson, T.D.; Snell, C.; Young, K.M.; Ernst, M.; Kilpatrick, T.J. SOCS3 negatively regulates LIF signaling in neural precursor cells. Mol. Cell Neurosci. 2006, 31, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Moriano, J.; Boeckx, C. Modern human changes in regulatory regions implicated in cortical development. BMC Genom. 2020, 21, 304. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Fos (TPM Values) | |||||||
|---|---|---|---|---|---|---|---|
| WT | Sox2-del | ||||||
| Mouse number | 1 | 2 | 3 | 1 | 2 | 3 | |
| D0 | 3430.60 | 2984.97 | 2189.25 | 978.08 | 259.14 | 1218.42 | |
| D4 | 14.52 | 4.38 | 7.48 | 5.28 | 4.92 | 4.81 | |
| D11 | 22.80 | 58.71 | 32.46 | 29.79 | 64.64 | 35.61 | |
| Socs3 (TPM Values) | |||||||
| WT | Sox2-del | ||||||
| Mouse number | 1 | 2 | 3 | 1 | 2 | 3 | |
| D0 | 235.80 | 336.16 | 191.33 | 27.50 | 11.18 | 49.28 | |
| D4 | 9.65 | 4.29 | 4.33 | 3.30 | 3.69 | 1.13 | |
| D11 | 20.20 | 19.08 | 21.81 | 41.68 | 50.67 | 44.26 | |
| Sox2 (TPM Values) | |||||||
| WT | Sox2-del | ||||||
| Mouse number | 1 | 2 | 3 | 1 | 2 | 3 | |
| D0 | 495.40 | 306.24 | 247.93 | 2.59 | 6.64 | 3.45 | |
| D4 | 332.73 | 151.27 | 307.37 | 5.00 | 21.53 | 8.02 | |
| D11 | 200.03 | 224.55 | 242.31 | 11.84 | 26.34 | 17.22 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pagin, M.; Pernebrink, M.; Pitasi, M.; Malighetti, F.; Ngan, C.-Y.; Ottolenghi, S.; Pavesi, G.; Cantù, C.; Nicolis, S.K. FOS Rescues Neuronal Differentiation of Sox2-Deleted Neural Stem Cells by Genome-Wide Regulation of Common SOX2 and AP1(FOS-JUN) Target Genes. Cells 2021, 10, 1757. https://doi.org/10.3390/cells10071757
Pagin M, Pernebrink M, Pitasi M, Malighetti F, Ngan C-Y, Ottolenghi S, Pavesi G, Cantù C, Nicolis SK. FOS Rescues Neuronal Differentiation of Sox2-Deleted Neural Stem Cells by Genome-Wide Regulation of Common SOX2 and AP1(FOS-JUN) Target Genes. Cells. 2021; 10(7):1757. https://doi.org/10.3390/cells10071757
Chicago/Turabian StylePagin, Miriam, Mattias Pernebrink, Mattia Pitasi, Federica Malighetti, Chew-Yee Ngan, Sergio Ottolenghi, Giulio Pavesi, Claudio Cantù, and Silvia K. Nicolis. 2021. "FOS Rescues Neuronal Differentiation of Sox2-Deleted Neural Stem Cells by Genome-Wide Regulation of Common SOX2 and AP1(FOS-JUN) Target Genes" Cells 10, no. 7: 1757. https://doi.org/10.3390/cells10071757
APA StylePagin, M., Pernebrink, M., Pitasi, M., Malighetti, F., Ngan, C. -Y., Ottolenghi, S., Pavesi, G., Cantù, C., & Nicolis, S. K. (2021). FOS Rescues Neuronal Differentiation of Sox2-Deleted Neural Stem Cells by Genome-Wide Regulation of Common SOX2 and AP1(FOS-JUN) Target Genes. Cells, 10(7), 1757. https://doi.org/10.3390/cells10071757