CytokineLink: A Cytokine Communication Map to Analyse Immune Responses—Case Studies in Inflammatory Bowel Disease and COVID-19
Abstract
:1. Introduction
2. Materials and Methods
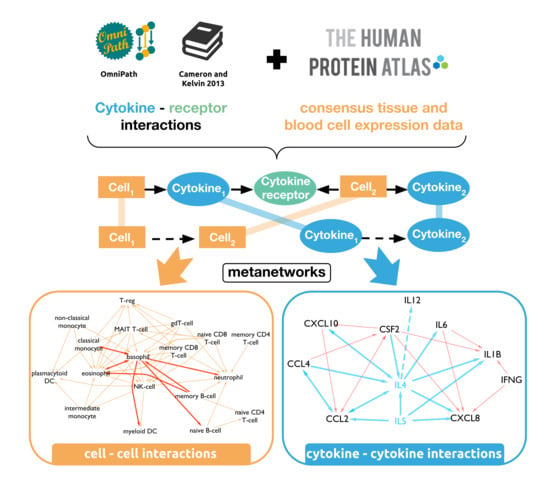
2.1. Network Generation
2.2. Annotation
2.3. Assessing the Translational Relevance of the Cytokine–Cytokine Interactions
2.4. Use Cases
3. Results
3.1. A Novel Network Resource of Cytokine Communication
3.2. Cytokine–Cytokine Meta-Edges Capture the Possible Ways Cytokines Affect Each Other
3.3. Cytokine Signalling in Ulcerative Colitis (UC)
3.4. Cytokine Signalling following SARS-CoV-2 Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cameron, M.J.; Kelvin, D.J. Cytokines and Chemokines-Their Receptors and Their Genes: An Overview. Madame Curie Bioscience Database; Landes Bioscience: Austin, TX, USA, 2003. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6294/ (accessed on 3 May 2020).
- Frankenstein, Z.; Alon, U.; Cohen, I.R. The Immune-Body Cytokine Network Defines a Social Architecture of Cell Interactions. Biol. Direct 2006, 1, 32. [Google Scholar] [CrossRef] [Green Version]
- Ozaki, K.; Leonard, W.J. Cytokine and Cytokine Receptor Pleiotropy and Redundancy. J. Biol. Chem. 2002, 277, 29355–29358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Q.; Wang, B.; Mao, J. The Pathogenesis and Treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and Tumor Progression: Signaling Pathways and Targeted Intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef] [PubMed]
- Tisoncik, J.R.; Korth, M.J.; Simmons, C.P.; Farrar, J.; Martin, T.R.; Katze, M.G. Into the Eye of the Cytokine Storm. Microbiol. Mol. Biol. Rev. 2012, 76, 16–32. [Google Scholar] [CrossRef] [Green Version]
- Costela-Ruiz, V.J.; Illescas-Montes, R.; Puerta-Puerta, J.M.; Ruiz, C.; Melguizo-Rodríguez, L. SARS-CoV-2 Infection: The Role of Cytokines in COVID-19 Disease. Cytokine Growth Factor Rev. 2020, 54, 62–75. [Google Scholar] [CrossRef]
- Altan-Bonnet, G.; Mukherjee, R. Cytokine-Mediated Communication: A Quantitative Appraisal of Immune Complexity. Nat. Rev. Immunol. 2019, 19, 205–217. [Google Scholar] [CrossRef]
- Dhillon, B.K.; Smith, M.; Baghela, A.; Lee, A.H.Y.; Hancock, R.E.W. Systems Biology Approaches to Understanding the Human Immune System. Front. Immunol. 2020, 11, 1683. [Google Scholar] [CrossRef]
- Türei, D.; Valdeolivas, A.; Gul, L.; Palacio-Escat, N.; Klein, M.; Ivanova, O.; Ölbei, M.; Gábor, A.; Theis, F.; Módos, D.; et al. Integrated Intra- and Intercellular Signaling Knowledge for Multicellular Omics Analysis. Mol. Syst. Biol. 2021, 17, e9923. [Google Scholar] [CrossRef] [PubMed]
- Atallah, M.B.; Tandon, V.; Hiam, K.J.; Boyce, H.; Hori, M.; Atallah, W.; Spitzer, M.H.; Engleman, E.; Mallick, P. ImmunoGlobe: Enabling Systems Immunology with a Manually Curated Intercellular Immune Interaction Network. BMC Bioinform. 2020, 21, 346. [Google Scholar] [CrossRef]
- Kveler, K.; Starosvetsky, E.; Ziv-Kenet, A.; Kalugny, Y.; Gorelik, Y.; Shalev-Malul, G.; Aizenbud-Reshef, N.; Dubovik, T.; Briller, M.; Campbell, J.; et al. Immune-Centric Network of Cytokines and Cells in Disease Context Identified by Computational Mining of PubMed. Nat. Biotechnol. 2018, 36, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Arijs, I.; De Hertogh, G.; Lemaire, K.; Quintens, R.; Van Lommel, L.; Van Steen, K.; Leemans, P.; Cleynen, I.; Van Assche, G.; Vermeire, S.; et al. Mucosal Gene Expression of Antimicrobial Peptides in Inflammatory Bowel Disease before and after First Infliximab Treatment. PLoS ONE 2009, 4, e7984. [Google Scholar] [CrossRef] [PubMed]
- Tasaki, S.; Suzuki, K.; Kassai, Y.; Takeshita, M.; Murota, A.; Kondo, Y.; Ando, T.; Nakayama, Y.; Okuzono, Y.; Takiguchi, M.; et al. Multi-Omics Monitoring of Drug Response in Rheumatoid Arthritis in Pursuit of Molecular Remission. Nat. Commun. 2018, 9, 2755. [Google Scholar] [CrossRef] [Green Version]
- Smillie, C.S.; Biton, M.; Ordovas-Montanes, J.; Sullivan, K.M.; Burgin, G.; Graham, D.B.; Herbst, R.H.; Rogel, N.; Slyper, M.; Waldman, J.; et al. Intra- and Inter-Cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 2019, 178, 714–730.e22. [Google Scholar] [CrossRef] [PubMed]
- Beal, J. Biochemical Complexity Drives Log-Normal Variation in Genetic Expression. Eng. Biol. 2017, 1, 55–60. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T. Ulcerative Colitis. Nat. Rev. Dis. Primers 2020, 6, 74. [Google Scholar] [CrossRef]
- Cader, M.Z.; Kaser, A. Recent Advances in Inflammatory Bowel Disease: Mucosal Immune Cells in Intestinal Inflammation. Gut 2013, 62, 1653–1664. [Google Scholar] [CrossRef]
- West, N.R.; Hegazy, A.N.; Owens, B.M.J.; Bullers, S.J.; Linggi, B.; Buonocore, S.; Coccia, M.; Görtz, D.; This, S.; Stockenhuber, K.; et al. Oncostatin M Drives Intestinal Inflammation and Predicts Response to Tumor Necrosis Factor-Neutralizing Therapy in Patients with Inflammatory Bowel Disease. Nat. Med. 2017, 23, 579–589. [Google Scholar] [CrossRef]
- Olbei, M.; Hautefort, I.; Modos, D.; Treveil, A.; Poletti, M.; Gul, L.; Shannon-Lowe, C.D.; Korcsmaros, T. SARS-CoV-2 Causes a Different Cytokine Response Compared to Other Cytokine Storm-Causing Respiratory Viruses in Severely Ill Patients. Front. Immunol. 2021, 12, 629193. [Google Scholar] [CrossRef]
- Thul, P.J.; Lindskog, C. The Human Protein Atlas: A Spatial Map of the Human Proteome. Protein Sci. 2018, 27, 233–244. [Google Scholar] [CrossRef] [Green Version]
- El-Kady, I.M.; El-Masry, S.A. Pro-Inflammatory and Anti-Inflammatory Cytokines Profile in Rheumatoid Arthritis Patients. Egypt. J. Immunol. 2008, 15, 109–114. [Google Scholar]
- Sanchez-Muñoz, F. Role of Cytokines in Inflammatory Bowel Disease. World J. Gastroenterol. WJG 2008, 14, 4280. [Google Scholar] [CrossRef] [PubMed]
- Ungaro, R.; Mehandru, S.; Allen, P.B.; Peyrin-Biroulet, L.; Colombel, J.-F. Ulcerative Colitis. Lancet 2017, 389, 1756–1770. [Google Scholar] [CrossRef]
- GBD 2017 Inflammatory Bowel Disease Collaborators. The Global, Regional, and National Burden of Inflammatory Bowel Disease in 195 Countries and Territories, 1990-2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17–30. [Google Scholar] [CrossRef] [Green Version]
- Gajendran, M.; Loganathan, P.; Jimenez, G.; Catinella, A.P.; Ng, N.; Umapathy, C.; Ziade, N.; Hashash, J.G. A Comprehensive Review and Update on Ulcerative Colitis. Dis. Mon. 2019, 65, 100851. [Google Scholar] [CrossRef]
- Kayal, M.; Shah, S. Ulcerative Colitis: Current and Emerging Treatment Strategies. J. Clin. Med. 2019, 9, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, E.L.; Jiang, Y.; Kappelman, M.D.; Long, M.D.; Sandler, R.S.; Kinlaw, A.C.; Herfarth, H.H. Decreasing Colectomy Rate for Ulcerative Colitis in the United States between 2007 and 2016: A Time Trend Analysis. Inflamm. Bowel Dis. 2020, 26, 1225–1231. [Google Scholar] [CrossRef] [PubMed]
- Privitera, G.; Pugliese, D.; Lopetuso, L.R.; Scaldaferri, F.; Neri, M.; Guidi, L.; Gasbarrini, A.; Armuzzi, A. Novel Trends with Biologics in Inflammatory Bowel Disease: Sequential and Combined Approaches. Ther. Adv. Gastroenterol. 2021, 14, 17562848211006668. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, C.; Cao, Y.; Rundquist, S.; Zhulina, Y.; Henriksson, I.; Montgomery, S.; Halfvarson, J. Changes in Medical Management and Colectomy Rates: A Population-Based Cohort Study on the Epidemiology and Natural History of Ulcerative Colitis in Örebro, Sweden, 1963-2010. Aliment. Pharmacol. Ther. 2017, 46, 748–757. [Google Scholar] [CrossRef] [Green Version]
- Denson, L.A.; Curran, M.; McGovern, D.P.B.; Koltun, W.A.; Duerr, R.H.; Kim, S.C.; Sartor, R.B.; Sylvester, F.A.; Abraham, C.; de Zoeten, E.F.; et al. Challenges in IBD Research: Precision Medicine. Inflamm. Bowel Dis. 2019, 25, S31–S39. [Google Scholar] [CrossRef] [Green Version]
- Kinchen, J.; Chen, H.H.; Parikh, K.; Antanaviciute, A.; Jagielowicz, M.; Fawkner-Corbett, D.; Ashley, N.; Cubitt, L.; Mellado-Gomez, E.; Attar, M.; et al. Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell 2018, 175, 372–386.e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, J.C.; Chang, C.; Boschetti, G.; Ungaro, R.; Giri, M.; Grout, J.A.; Gettler, K.; Chuang, L.-S.; Nayar, S.; Greenstein, A.J.; et al. Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 2019, 178, 1493–1508.e20. [Google Scholar] [CrossRef]
- Parikh, K.; Antanaviciute, A.; Fawkner-Corbett, D.; Jagielowicz, M.; Aulicino, A.; Lagerholm, C.; Davis, S.; Kinchen, J.; Chen, H.H.; Alham, N.K.; et al. Colonic Epithelial Cell Diversity in Health and Inflammatory Bowel Disease. Nature 2019, 567, 49–55. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; HLH Across Speciality Collaboration, UK. COVID-19: Consider Cytokine Storm Syndromes and Immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Shimabukuro-Vornhagen, A.; Gödel, P.; Subklewe, M.; Stemmler, H.J.; Schlößer, H.A.; Schlaak, M.; Kochanek, M.; Böll, B.; von Bergwelt-Baildon, M.S. Cytokine Release Syndrome. J. Immunother. Cancer 2018, 6, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tjan, L.H.; Furukawa, K.; Nagano, T.; Kiriu, T.; Nishimura, M.; Arii, J.; Hino, Y.; Iwata, S.; Nishimura, Y.; Mori, Y. Early Differences in Cytokine Production by Severity of Coronavirus Disease 2019. J. Infect. Dis. 2021, 223, 1145–1149. [Google Scholar] [CrossRef]
- Merza, M.Y.; Hwaiz, R.A.; Hamad, B.K.; Mohammad, K.A.; Hama, H.A.; Karim, A.Y. Analysis of Cytokines in SARS-CoV-2 or COVID-19 Patients in Erbil City, Kurdistan Region of Iraq. PLoS ONE 2021, 16, e0250330. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.-S.; Wang, X.-R.; Zhang, J.-C.; Yang, W.-B.; Ma, W.-L.; Yang, B.-H.; Jiang, N.-C.; Gao, Z.-C.; Shi, H.-Z.; Zhou, Q. A Cluster of Health Care Workers with COVID-19 Pneumonia Caused by SARS-CoV-2. J. Microbiol. Immunol. Infect. 2020. [Google Scholar] [CrossRef] [PubMed]
- Seder, R.A.; Paul, W.E. Acquisition of Lymphokine-Producing Phenotype by CD4 + T Cells. Annu. Rev. Immunol. 1994, 12, 635–673. [Google Scholar] [CrossRef]
- Kang, S.; Brown, H.M.; Hwang, S. Direct Antiviral Mechanisms of Interferon-Gamma. Immune Netw. 2018, 18, e33. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Ashkar, A.A. The Dual Nature of Type I and Type II Interferons. Front. Immunol. 2018, 9, 2061. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, N.; Seder, R.A.; Reddy, A.; Feldman, M.V. IL-12 in Conjunction with Dendritic Cells Enhances Antiviral CD8 + CTL Responses in Vitro. J. Clin. Investig. 1996, 98, 715–722. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.-Y.; Wang, S.-H.; Wu, C.-H.; Yen, L.-C.; Lai, H.-F.; Ho, C.-L.; Chiu, Y.-L. In Silico Immune Infiltration Profiling Combined with Functional Enrichment Analysis Reveals a Potential Role for Naïve B Cells as a Trigger for Severe Immune Responses in the Lungs of COVID-19 Patients. PLoS ONE 2020, 15, e0242900. [Google Scholar] [CrossRef] [PubMed]
- Pratt, D.; Chen, J.; Pillich, R.; Rynkov, V.; Gary, A.; Demchak, B.; Ideker, T. Ndex 2.0: A Clearinghouse for Research on Cancer Pathways. Cancer Res. 2017, 77, e58–e61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, M.D.; Nedjai, B.; Hurst, T.; Pennington, D.J. Cytokines and Chemokines: At the Crossroads of Cell Signalling and Inflammatory Disease. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 2563–2582. [Google Scholar] [CrossRef] [Green Version]
- Kany, S.; Vollrath, J.T.; Relja, B. Cytokines in Inflammatory Disease. Int. J. Mol. Sci. 2019, 20, 6008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grün, D.; Lyubimova, A.; Kester, L.; Wiebrands, K.; Basak, O.; Sasaki, N.; Clevers, H.; van Oudenaarden, A. Single-Cell Messenger RNA Sequencing Reveals Rare Intestinal Cell Types. Nature 2015, 525, 251–255. [Google Scholar] [CrossRef]
- Ramilowski, J.A.; Goldberg, T.; Harshbarger, J.; Kloppmann, E.; Lizio, M.; Satagopam, V.P.; Itoh, M.; Kawaji, H.; Carninci, P.; Rost, B.; et al. A Draft Network of Ligand-Receptor-Mediated Multicellular Signalling in Human. Nat. Commun. 2015, 6, 7866. [Google Scholar] [CrossRef] [Green Version]
- Batista, F.D.; Dustin, M.L. Cell: Cell Interactions in the Immune System. Immunol. Rev. 2013, 251, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-Cell Landscape of Bronchoalveolar Immune Cells in Patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Daneshpour, H.; Youk, H. Modeling Cell-Cell Communication for Immune Systems across Space and Time. Curr. Opin. Syst. Biol. 2019, 18, 44–52. [Google Scholar] [CrossRef] [PubMed]



| Dataset | Reference | Drug | Inhibited Cytokine | p-Value |
|---|---|---|---|---|
| GSE16879 | [13] | infliximab | TNFα | 0.01504 |
| GSE92415 | (Li et al., 2018, unpublished data) | golimumab | TNFα | 0.01525 |
| GSE93777 | [14] | tocilizumab | IL6 | 0.00587 |
| Tissues | 24 | |
| Blood cell types | 18 | |
| Cytokines | 115 | |
| Cytokine–receptor pairs | 260 | |
| Interactions | ||
| cell–cell | All interactions between two cells regardless of receptor usage | 581 |
| All interactions listed between two cells, mediated by different receptors | 1118 | |
| cytokine–cytokine | All interactions between two cytokines regardless of receptor usage | 2818 |
| All interactions listed between two cytokines, mediated by different receptors | 9195 | |
| Annotated interactions | ||
| cell–cell | Number of cell–cell interactions with annotated cell–cytokine relationships | 74 |
| Percentage of cell–cell interactions with annotated cell–cytokine relationships | 6.7% | |
| cytokine–cytokine | Number of cytokine–cytokine interactions with annotated cell–cytokine relationships | 1673 |
| Percentage of cytokine–cytokine interactions with annotated cell–cytokine relationships | 18.2% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olbei, M.; Thomas, J.P.; Hautefort, I.; Treveil, A.; Bohar, B.; Madgwick, M.; Gul, L.; Csabai, L.; Modos, D.; Korcsmaros, T. CytokineLink: A Cytokine Communication Map to Analyse Immune Responses—Case Studies in Inflammatory Bowel Disease and COVID-19. Cells 2021, 10, 2242. https://doi.org/10.3390/cells10092242
Olbei M, Thomas JP, Hautefort I, Treveil A, Bohar B, Madgwick M, Gul L, Csabai L, Modos D, Korcsmaros T. CytokineLink: A Cytokine Communication Map to Analyse Immune Responses—Case Studies in Inflammatory Bowel Disease and COVID-19. Cells. 2021; 10(9):2242. https://doi.org/10.3390/cells10092242
Chicago/Turabian StyleOlbei, Marton, John P. Thomas, Isabelle Hautefort, Agatha Treveil, Balazs Bohar, Matthew Madgwick, Lejla Gul, Luca Csabai, Dezso Modos, and Tamas Korcsmaros. 2021. "CytokineLink: A Cytokine Communication Map to Analyse Immune Responses—Case Studies in Inflammatory Bowel Disease and COVID-19" Cells 10, no. 9: 2242. https://doi.org/10.3390/cells10092242
APA StyleOlbei, M., Thomas, J. P., Hautefort, I., Treveil, A., Bohar, B., Madgwick, M., Gul, L., Csabai, L., Modos, D., & Korcsmaros, T. (2021). CytokineLink: A Cytokine Communication Map to Analyse Immune Responses—Case Studies in Inflammatory Bowel Disease and COVID-19. Cells, 10(9), 2242. https://doi.org/10.3390/cells10092242