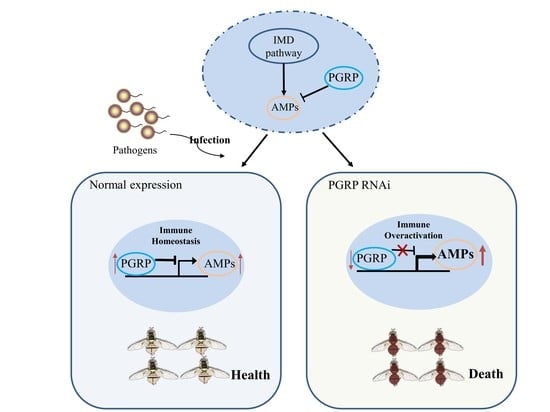
The Negative Regulative Roles of BdPGRPs in the Imd Signaling Pathway of Bactrocera dorsalis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Insects
2.2. Cloning and Analysis of the BdPGRP Genes
2.3. Development Stage and Tissue Expression Profiles
2.4. Bacterial Preparation and Infection Bioassays
2.5. Double Strain RNA Synthesis and RNAi
2.6. Investigation of the RNAi Off-Target Effect and RNAi Efficiency
2.7. The Effects of Knockdown of BdPGRPs on the Imd Pathway Response to Bacterial Challenge
2.8. Survival Assay of B. dorsalis
2.9. Quantitative Real-Time PCR
2.10. Statistical Analyses
3. Results
3.1. Sequence Features, Phylogenetic Tree and Functional Domain Prediction of PGRPs in B. dorsalis
3.2. The Expression Profilings of BdPGRPs in B. dorsalis
3.3. Responses of BdPGRPs to Systemic Bacterial Infection
3.4. RNA Interference (RNAi) of BdPGRPs
3.5. The Negative Regulatory Roles of BdPGRPs in Imd Pathway
3.6. BdPGRPs RNAi Decreased Flies Survival Rate after Bacterial Challenge
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tzou, P.; De Gregorio, E.; Lemaitre, B. How Drosophila combats microbial infection: A model to study innate immunity and host-pathogen interactions. Curr. Opin. Microbiol. 2002, 5, 102–110. [Google Scholar] [CrossRef]
- Janeway, C.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef] [Green Version]
- Guan, R.; Mariuzza, R.A. Peptidoglycan recognition proteins of the innate immune system. Trends Microbiol. 2007, 15, 127–134. [Google Scholar] [CrossRef]
- Royet, J.; Dziarski, R. Peptidoglycan recognition proteins: Pleiotropic sensors and effectors of antimicrobial defences. Nat. Rev. Microbiol. 2007, 5, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Mengin-Lecreulx, D.; Lemaitre, B. Structure and metabolism of peptidoglycan and molecular requirements allowing its detection by the Drosophila innate immune system. J. Endotoxin Res. 2005, 11, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Lemaitre, B.; Hoffmann, J. The Host Defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, H.; Kinoshita, K.; Ashida, M. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J. Biol. Chem. 1996, 271, 13854–13860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, T.; Liu, G.; Kang, D.; Ekengren, S.; Steiner, H.; Hultmark, D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2000, 97, 13772–13777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christophides, G.; Zdobnov, E.; Barillas-Mury, C.; Birney, E.; Blandin, S.; Blass, C.; Brey, P.; Collins, F.; Danielli, A.; Dimopoulos, G.; et al. Immunity-related genes and gene families in Anopheles gambiae. Science 2002, 298, 159–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dziarski, R.; Gupta, D. The peptidoglycan recognition proteins (PGRPs). Genome Biol. 2006, 7, 232. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Beerntsen, B.T. Insights into the different functions of multiple peptidoglycan recognition proteins in the immune response against bacteria in the mosquito, Armigeres subalbatus. Insect Biochem. Mol. Biol. 2013, 43, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Buchon, N.; Broderick, N.A.; Lemaitre, B. Gut homeostasis in a microbial world: Insights from Drosophila melanogaster. Nat. Rev. Microbiol. 2013, 11, 615–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paredes, J.C.; Welchman, D.P.; Poidevin, M.; Lemaitre, B. Negative Regulation by Amidase PGRPs Shapes the Drosophila Antibacterial Response and Protects the Fly from Innocuous Infection. Immunity. 2011, 35, 770–779. [Google Scholar] [CrossRef] [Green Version]
- Mellroth, P.; Karlsson, J.; Steiner, H. A scavenger function for a Drosophila peptidoglycan recognition protein. J. Biol. Chem. 2003, 278, 7059–7064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Karpac, J.; Tran, S.L.; Jasper, H. PGRP-SC2 Promotes Gut Immune Homeostasis to Limit Commensal Dysbiosis and Extend Lifespan. Cell 2014, 156, 109–122. [Google Scholar] [CrossRef] [Green Version]
- Zaidman-Rémy, A.; Poidevin, M.; Hervé, M.; Welchman, D.P.; Paredes, J.C.; Fahlander, C.; Steiner, H.; Mengin-Lecreulx, D.; Lemaitre, B. Drosophila Immunity: Analysis of PGRP-SB1 Expression, Enzymatic Activity and Function. PLoS ONE 2011, 6, e17231. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, M.; Zhang, H. RNA interference of four genes in adult Bactrocera dorsalis by feeding their dsRNAs. PLoS ONE 2011, 6, e17788. [Google Scholar] [CrossRef]
- Clarke, A.R.; Armstrong, K.F.; Carmichael, A.E.; Milne, J.R.; Raghu, S.; Roderick, G.K.; Yeates, D.K. Invasive phytophagous pests arising through a recent tropical evolutionary radiation: The Bactrocera dorsalis Complex of Fruit Flies. Annu. Rev. Entomol. 2005, 50, 293–319. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Dong, X.; Zheng, W.; Zhang, H. The PLA2 gene mediates the humoral immune responses in Bactrocera dorsalis (Hendel). Dev. Comp. Immunol. 2017, 67, 293–299. [Google Scholar] [CrossRef]
- Dong, X.; Li, Q.; Zhang, H. The noa gene is functionally linked to the activation of the Toll/Imd signaling pathways in Bactrocera dorsalis (Hendel). Dev. Comp. Immunol. 2016, 55, 233–240. [Google Scholar] [CrossRef]
- Yao, Z.; Wang, A.; Li, Y.; Cai, Z.; Lemaitre, B.; Zhang, H. The dual oxidase gene BdDuox regulates the intestinal bacterial community homeostasis of Bactrocera dorsalis. ISME J. 2016, 10, 1037–1050. [Google Scholar] [CrossRef] [Green Version]
- Gottar, M.; Gobert, V.; Michel, T.; Belvin, M.; Duyk, G.; Hoffmann, J.; Ferrandon, D.; Royet, J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 2002, 416, 640–644. [Google Scholar] [CrossRef]
- Bischoff, V.; Vignal, C.; Boneca, I.G.; Michel, T.; Hoffmann, J.A.; Royet, J. Function of the Drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat. Immunol. 2004, 5, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Yano, T.; Aggarwal, K.; Lim, J.H.; Ueda, K.; Oshima, Y.; Peach, C.; Erturk-Hasdemir, D.; Goldman, W.E.; Oh, B.H.; et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 2006, 7, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Kurata, S. Extracellular and intracellular pathogen recognition by Drosophila PGRP-LE and PGRP-LC. Int. Immunol. 2010, 22, 143–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Tang, T.; Gu, J.; Sun, L.; Gao, X.; Ma, X.; Wang, X.; Liu, F.; Wang, J. Downregulation of the Musca domestica peptidoglycan recognition protein SC (PGRP-SC) leads to overexpression of antimicrobial peptides and tardy pupation. Mol. Immunol. 2015, 67, 465–474. [Google Scholar] [CrossRef]
- Dawadi, B.; Wang, X.; Xiao, R.; Muhammad, A.; Hou, Y.; Shi, Z. PGRP-LB homolog acts as a negative modulator of immunity in maintaining the gut-microbe symbiosis of red palm weevil, Rhynchophorus ferrugineus Olivier. Dev. Comp. Immunol. 2018, 86, 65–77. [Google Scholar] [CrossRef]
- Maire, J.; Vincent-Monégat, C.; Balmand, S.; Vallier, A.; Hervé, M.; Masson, F.; Parisot, N.; Vigneron, A.; Anselme, C.; Perrin, J.; et al. Weevil pgrp-lb prevents endosymbiont TCT dissemination and chronic host systemic immune activation. Proc. Natl. Acad. Sci. USA 2019, 116, 5623–5632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leulier, F.; Parquet, C.; Pili-Floury, S.; Ryu, J.; Caroff, M.; Lee, W.; Mengin-Lecreulx, D.; Lemaitre, B. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 2003, 4, 478–484. [Google Scholar] [CrossRef]
- Seinen, E.; Burgerhof, J.G.M.; Jansen, R.C.; Sibon, O.C.M. RNAi-induced off-target effects in Drosophila melanogaster: Frequencies and solutions. Brief. Funct. Genom. 2011, 10, 206–214. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lim, J.H.; Kim, M.S.; Kim, H.E.; Yano, T.; Oshima, Y.; Aggarwal, K.; Goldman, W.E.; Silverman, N.; Kurata, S.; Oh, B.H. Structural Basis for Preferential Recognition of Diaminopimelic Acid-type Peptidoglycan by a Subset of Peptidoglycan Recognition Proteins. J. Biol. Chem. 2006, 281, 8286–8295. [Google Scholar] [CrossRef] [Green Version]
- Beyenbach, K.; Skaer, H.; Dow, J. The developmental, molecular, and transport biology of Malpighian tubules. Annu. Rev. Entomol. 2010, 55, 351–374. [Google Scholar] [CrossRef]
- Kurata, S. Recognition of infectious non-self and activation of immune responses by peptidoglycan recognition protein (PGRP)-family members in Drosophila. Dev. Comp. Immunol. 2004, 28, 89–95. [Google Scholar] [CrossRef]
- Ferrandon, D.; Imler, J.; Hetru, C.; Hoffmann, J. The Drosophila systemic immune response: Sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 2007, 7, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Qi, J.; Wang, S.; Wang, M.; Li, X.; Kim, Y.G.; Núñez, G.; Gupta, D.; Dziarski, R. PGLYRP-2 and Nod2 are both required for peptidoglycan-induced arthritis and local inflammation. Cell Host Microbe 2009, 5, 137–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosco-Drayon, V.; Poidevin, M.; Boneca, I.G.; Narbonne-Reveau, K.; Royet, J.; Charroux, B. Peptidoglycan Sensing by the Receptor PGRP-LE in the Drosophila Gut Induces Immune Responses to Infectious Bacteria and Tolerance to Microbiota. Cell Host Microbe 2012, 12, 153–165. [Google Scholar] [CrossRef] [Green Version]
- Tindwa, H.; Patnaik, B.B.; Kim, D.H.; Mun, S.; Jo, Y.H.; Lee, B.L.; Lee, Y.S.; Kim, N.J.; Han, Y.S. Cloning, characterization and effect of TmPGRP-LE gene silencing on survival of Tenebrio molitor against Listeria monocytogenes infection. Int. J. Mol. Sci. 2013, 14, 22462–22482. [Google Scholar] [CrossRef]
- Mendes, C.; Felix, R.; Sousa, A.M.; Lamego, J.; Charlwood, D.; do Rosário, V.E.; Pinto, J.; Silveira, H. Molecular evolution of the three short PGRPs of the malaria vectors Anopheles gambiae and Anopheles arabiensis in East Africa. BMC Evol. Biol. 2010, 10, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, Y.Y.; Qu, L.Y.; Zhao, D.; Chen, L.B.; Jin, H.Y.; Xu, L.M.; Cheng, J.A.; Zhang, C.X. The genome- and transcriptome-wide analysis of innate immunity in the brown planthopper, Nilaparvata lugens. BMC Genom. 2013, 14, 160. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Aksoy, S. PGRP-LB is a maternally transmitted immune milk protein that influences symbiosis and parasitism in tsetse’s offspring. Proc. Natl. Acad. Sci. USA 2012, 109, 10552–10557. [Google Scholar] [CrossRef] [Green Version]
- Tsakas, S.; Marmaras, V.J. Insect immunity and its signalling: An overview. Invertebr. Surviv. J. 2010, 7, 228–238. [Google Scholar]
- Li, S.; Yu, X.; Feng, Q. Fat Body Biology in the Last Decade. Annu. Rev. Entomol. 2019, 64, 315–333. [Google Scholar] [CrossRef] [PubMed]
- Zaidman-Rémy, A.; Hervé, M.; Poidevin, M.; Pili-Floury, S.; Kim, M.-S.; Blanot, D.; Oh, B.-H.; Ueda, R.; Mengin-Lecreulx, D.; Lemaitre, B. The Drosophila Amidase PGRP-LB Modulates the Immune Response to Bacterial Infection. Immunity 2006, 24, 463–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eijkelenboom, A.; Burgering, B.M. FOXOs: Signalling integrators for homeostasis maintenance. Nature Reviews Molecular Cell Biology 2013, 14, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.; Bischoff, V.; Vignal, C.; Duvic, B.; Boneca, I.G.; Hoffmann, J.A.; Royet, J. Downregulation of the Drosophila Immune Response by Peptidoglycan-Recognition Proteins SC1 and SC2. PLoS Pathog. 2006, 2, e14. [Google Scholar]
- Charroux, B.; Capo, F.; Kurz, C.L.; Peslier, S.; Chaduli, D.; Viallat-Lieutaud, A.; Royet, J. Cytosolic and Secreted Peptidoglycan-Degrading Enzymes in Drosophila Respectively Control Local and Systemic Immune Responses to Microbiota. Cell Host Microbe 2018, 23, 215–228. [Google Scholar] [CrossRef] [Green Version]
- Dziarski, R.; Gupta, D. A balancing act: PGRPs preserve and protect. Cell Host Microbe 2018, 23, 149–151. [Google Scholar] [CrossRef] [Green Version]
- Dolezal, T.; Krejcova, G.; Bajgar, A.; Nedbalova, P.; Strasser, P. Molecular regulations of metabolism during immune response in insects. Insect Biochem. Mol. Biol. 2019, 109, 31–42. [Google Scholar] [CrossRef]







| Primer | Sequence (from 5′ to 3′) | Purpose |
|---|---|---|
| PGRP-SC2 5′RACE outer | CCTTAGCGGCAGCAATCT | RACE |
| PGRP-SC2 5′RACE inner | CCACGACCCTCATACACT | RACE |
| PGRP-SC2 3′RACE outer | GCAAGTGTATGAGGGTCG | RACE |
| PGRP-SC2 3′RACE inner | TTACTGCTCCACCCAAAC | RACE |
| QPGRP-LB F | GCGTGGCTGGAATGACATTG | qRT-PCR |
| QPGRP-LB R | CGGTCATTGTATTTGGGCGC | qRT-PCR |
| QPGRP-SB F | TGGCATTGTCTTCATCGGCA | qRT-PCR |
| QPGRP-SB R | CAGATAACCCTTTTGCACCGC | qRT-PCR |
| QPGRP-SC2 F | GGGTCGTGGTTGGAGTACAG | qRT-PCR |
| QPGRP-SC2 R | GATCTGAGCGGCTGTTGGAA | qRT-PCR |
| QRpL32 F | CCCGTCATATGCTGCCAACT | qRT-PCR |
| QRpL32 R | GCGCGCTCAACAATTTCCTT | qRT-PCR |
| QDiptericin F | GCATAGATTTGAGCCTTGACACAC | qRT-PCR |
| QDiptericin R | GCCATATCGTCCGCCCAAAT | qRT-PCR |
| PGRP-LB T7F | GGATCCTAATACGACTCACTATAGGATGCCCAGCGCCTGTTAC | dsRNA synthesis |
| PGRP-LB T7R | GGATCCTAATACGACTCACTATAGGTGCGGCCACGTCGTAATC | dsRNA synthesis |
| PGRP-SB T7 F | GGATCCTAATACGACTCACTATAGGTGTTTTGCGCTCAGGATCCA | dsRNA synthesis |
| PGRP-SB T7R | GGATCCTAATACGACTCACTATAGGTGGCCCAGCAGTGTGTAATT | dsRNA synthesis |
| PGRP-SC2 T7 F | GGATCCTAATACGACTCACTATAGGGGCTTTCAAGACTTTCCTC | dsRNA synthesis |
| PGRP-SC2 T7R | GGATCCTAATACGACTCACTATAGGAACCACGACCCTCATACAC | dsRNA synthesis |
| EGFP T7L | GGATCCTAATACGACTCACTATAGGACGTAAACGGCCACAAGTTC | dsRNA synthesis |
| EGFP T7R | GGATCCTAATACGACTCACTATAGGAAGTCGTGCTGCTTAATGTG | dsRNA synthesis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Yao, Z.; Bai, S.; Zhang, H. The Negative Regulative Roles of BdPGRPs in the Imd Signaling Pathway of Bactrocera dorsalis. Cells 2022, 11, 152. https://doi.org/10.3390/cells11010152
Zhang P, Yao Z, Bai S, Zhang H. The Negative Regulative Roles of BdPGRPs in the Imd Signaling Pathway of Bactrocera dorsalis. Cells. 2022; 11(1):152. https://doi.org/10.3390/cells11010152
Chicago/Turabian StyleZhang, Ping, Zhichao Yao, Shuai Bai, and Hongyu Zhang. 2022. "The Negative Regulative Roles of BdPGRPs in the Imd Signaling Pathway of Bactrocera dorsalis" Cells 11, no. 1: 152. https://doi.org/10.3390/cells11010152
APA StyleZhang, P., Yao, Z., Bai, S., & Zhang, H. (2022). The Negative Regulative Roles of BdPGRPs in the Imd Signaling Pathway of Bactrocera dorsalis. Cells, 11(1), 152. https://doi.org/10.3390/cells11010152