Regulation of Conidiogenesis in Aspergillus flavus
Abstract
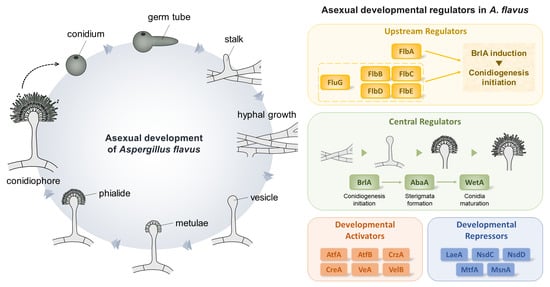
:1. Introduction
2. Distribution of Key Developmental Regulators in Aspergillus
2.1. Central Regulators of Conidiophore Production in Aspergillus
2.1.1. BrlA
2.1.2. AbaA
2.1.3. WetA
2.2. Upstream Regulators in Asexual Development
2.2.1. Developmental Activators
2.2.2. Velvet Regulators and LaeA
2.3. Other Key Regulators for Asexual Development
2.3.1. AtfA and AtfB
2.3.2. CreA
2.3.3. CrzA
2.3.4. HogA (SakA)
2.3.5. MsnA
2.3.6. MtfA
2.3.7. NsdC and NsdD
2.3.8. SfgA
2.3.9. StuA
3. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.-B.; Hubka, V.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.; Hubka, V.; Ezekiel, C.; Hong, S.-B.; Nováková, A.; Chen, A.; Arzanlou, M.; Larsen, T.; Sklenář, F.; Mahakarnchanakul, W.; et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 2018, 93, 1–63. [Google Scholar] [CrossRef] [PubMed]
- Amaike, S.; Keller, N.P. Aspergillus flavus . Annu. Rev. Phytopathol. 2011, 49, 107–133. [Google Scholar] [CrossRef]
- Klich, M.A. Aspergillus flavus: The major producer of aflatoxin. Mol. Plant Pathol. 2007, 8, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Manavathu, E.K.; Chandrasekar, P.H. Aspergillus flavus: An emerging non-fumigatus Aspergillus species of significance. Mycoses 2009, 52, 206–222. [Google Scholar] [CrossRef]
- Rudramurthy, S.M.; Paul, R.A.; Chakrabarti, A.; Mouton, J.W.; Meis, J.F. Invasive aspergillosis by Aspergillus flavus: Epidemiology, diagnosis, antifungal resistance, and management. J. Fungi 2019, 5, 55. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Jun, S.-C.; Han, K.-H.; Hong, S.-B.; Yu, J.-H. Diversity, application, and synthetic biology of industrially important Aspergillus fungi. Adv. Appl. Microbiol. 2017, 100, 161–202. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.-J.; Hu, S.; Wang, B.-T.; Jin, L. Advances in genetic engineering technology and its application in the industrial fungus Aspergillus oryzae. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Abe, K.; Asai, K.; Gomi, K.; Juvvadi, P.R.; Kato, M.; Kitamoto, K.; Takeuchi, M.; Machida, M. Genomics of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2007, 71, 646–670. [Google Scholar] [CrossRef] [PubMed]
- Batomunkueva, B.P.; Egorov, N.S. Preparations of extracellular proteinases from Aspergillus ochraceus 513 and Aspergillus al-liaceus 7dN1. Mikrobiologiia 2002, 71, 56–58. [Google Scholar]
- Varga, J.; Frisvad, J.; Samson, R. Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Stud. Mycol. 2011, 69, 57–80. [Google Scholar] [CrossRef]
- Bennett, J.W. An Overview of the Genus Aspergillus. In The Aspergillus; Molecular Biology and Genomics; CRC Press: Boca Raton, FL, USA, 2010; pp. 1–17. [Google Scholar]
- Adams, T.H.; Wieser, J.K.; Yu, J.-H. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 1998, 62, 35–54. [Google Scholar] [CrossRef] [PubMed]
- Ebbole, D.J. The Conidium. In Cellular and Molecular Biology of Filamentous Fungi; American Society for Microbiology Press: Washington, DC, USA, 2010; pp. 577–590. [Google Scholar]
- Baltussen, T.J.H.; Zoll, J.; Verweij, P.E.; Melchers, W.J.G. Molecular mechanisms of conidial germination in Aspergillus spp. Microbiol. Mol. Biol. Rev. 2020, 84, e00049-19. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Yu, J.-H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 2012, 15, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Blachowicz, A.; Raffa, N.; Bok, J.W.; Choera, T.; Knox, B.; Lim, F.Y.; Huttenlocher, A.; Wang, C.C.C.; Venkateswaran, K.; Keller, N.P. Contributions of spore secondary metabolites to UV-C protection and virulence vary in different Aspergillus fumigatus strains. mBio 2020, 11, e03415-19. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, H.H.; Ramaswamy, A.; Sim, S.C.; Keller, N.P. Increased conidiation associated with progression along the sterig-matocystin biosynthetic pathway. Mycologia 2004, 96, 1190–1198. [Google Scholar] [CrossRef]
- Ojeda-López, M.; Chen, W.; Eagle, C.; Gutiérrez, G.; Jia, W.; Swilaiman, S.; Huang, Z.; Park, H.-S.; Yu, J.-H.; Cánovas, D.; et al. Evolution of asexual and sexual reproduction in the aspergilli. Stud. Mycol. 2018, 91, 37–59. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Lee, M.-K.; Han, K.-H.; Kim, M.-J.; Yu, J.-H. Developmental decisions in Aspergillus nidulans. Biol. Fungal Cell 2019, 63–80. [Google Scholar] [CrossRef]
- Casselton, L.A.; Zolan, M.E. The art and design of genetic screens: Filamentous fungi. Nat. Rev. Genet. 2002, 3, 683–697. [Google Scholar] [CrossRef] [PubMed]
- De Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K.; et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [CrossRef] [PubMed]
- Kjærbølling, I.; Vesth, T.; Frisvad, J.C.; Nybo, J.L.; Theobald, S.; Kildgaard, S.; Petersen, T.I.; Kuo, A.; Sato, A.; Lyhne, E.K.; et al. A comparative genomics study of 23 Aspergillus species from section Flavi. Nat. Commun. 2020, 11, 1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyer, P.S.; O’Gorman, C.M. Sexual development and cryptic sexuality in fungi: Insights from Aspergillus species. FEMS Microbiol. Rev. 2012, 36, 165–192. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Frisvad, J.; Sun, B.; Varga, J.; Kocsubé, S.; Dijksterhuis, J.; Kim, D.; Hong, S.-B.; Houbraken, J.; Samson, R. Aspergillus section Nidulantes (formerly Emericella): Polyphasic taxonomy, chemistry and biology. Stud. Mycol. 2016, 84, 1–118. [Google Scholar] [CrossRef] [PubMed]
- Affeldt, K.J.; Carrig, J.; Amare, M.; Keller, N.P. Global survey of canonical Aspergillus flavus G protein-coupled receptors. mBio 2014, 5, e01501–14. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.K.; Mack, B.M.; Moore, G.G.; Downey, D.L.; Lebar, M.; Joardar, V.; Losada, L.; Yu, J.; Nierman, W.C.; Bhatnagar, D. Whole genome comparison of Aspergillus flavus L-morphotype strain NRRL 3357 (type) and S-morphotype strain AF70. PLoS ONE 2018, 13, e0199169. [Google Scholar] [CrossRef]
- Etxebeste, O.; Otamendi, A.; Garzia, A.; Espeso, E.A.; Cortese, M.S. Rewiring of transcriptional networks as a major event leading to the diversity of asexual multicellularity in fungi. Crit. Rev. Microbiol. 2019, 45, 548–563. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.I.; Lange, L.; Lacey, A.E.; Vuong, D.; Midgley, D.J.; Greenfield, P.; Bradbury, M.; Lacey, E.; Busk, P.K.; Pilgaard, B.; et al. Aspergillus hancockii sp. nov., a biosynthetically talented fungus endemic to southeastern Australian soils. PLoS ONE 2017, 12, e0170254. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.U.; Kim, K.M.; Choi, Y.-H.; Hurh, B.-S.; Lee, I. Whole genome analysis of Aspergillus sojae SMF 134 supports its merits as a starter for soybean fermentation. J. Microbiol. 2019, 57, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Pei, H.; Zhou, X.; Zhao, K.; Yu, M.; Han, G.; Fan, J.; Tao, F. Systematic characterization of bZIP transcription factors required for development and aflatoxin generation by high-throughput gene knockout in Aspergillus flavus. J. Fungi 2022, 8, 356. [Google Scholar] [CrossRef] [PubMed]
- Fasoyin, O.E.; Wang, B.; Qiu, M.; Han, X.; Chung, K.-R.; Wang, S. Carbon catabolite repression gene creA regulates morphology, aflatoxin biosynthesis and virulence in Aspergillus flavus. Fungal Genet. Biol. 2018, 115, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.-Y.; Son, Y.-E.; Lee, D.-H.; Eom, T.-J.; Kim, M.-J.; Park, H.-S. Function of crzA in fungal development and aflatoxin production in Aspergillus flavus. Toxins 2019, 11, 567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, R.; Yang, K.; Tumukunde, E.; Guo, Z.; Zhang, B.; Liu, Y.; Zhuang, Z.; Yuan, J.; Wang, S. Regulator of G protein signaling contributes to the development and aflatoxin biosynthesis in Aspergillus flavus through the regulation of Gα Activity. Appl. Environ. Microbiol. 2022, 88, e00244-22. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.-K.; Scharfenstein, L.L.; Mack, B.; Ehrlich, K.C. Deletion of the Aspergillus flavus orthologue of A. nidulans fluG reduces conidiation and promotes production of sclerotia but does not abolish aflatoxin biosynthesis. Appl. Environ. Microbiol. 2012, 78, 7557–7563. [Google Scholar] [CrossRef] [PubMed]
- Tumukunde, E.; Li, D.; Qin, L.; Li, Y.; Shen, J.; Wang, S.; Yuan, J. Osmotic-adaptation response of sakA/hogA gene to aflatoxin biosynthesis, morphology development and pathogenicity in Aspergillus flavus. Toxins 2019, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Amaike, S.; Keller, N.P. Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryot. Cell 2009, 8, 1051–1060. [Google Scholar] [CrossRef]
- Chang, P.-K.; Scharfenstein, L.L.; Luo, M.; Mahoney, N.; Molyneux, R.J.; Yu, J.; Brown, R.L.; Campbell, B.C. Loss of msnA, a putative stress regulatory gene, in Aspergillus parasiticus and Aspergillus flavus increased production of conidia, aflatoxins and kojic acid. Toxins 2011, 3, 82–104. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Lohmar, J.M.; Satterlee, T.; Cary, J.W.; Calvo, A.M. The master transcription factor mtfA governs aflatoxin production, morphological development and pathogenicity in the fungus Aspergillus flavus. Toxins 2016, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Cary, J.W.; Harris-Coward, P.Y.; Ehrlich, K.C.; Mack, B.M.; Kale, S.P.; Larey, C.; Calvo, A.M. NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot. Cell 2012, 11, 1104–1111. [Google Scholar] [CrossRef]
- Yuan, X.-Y.; Li, J.-Y.; Zhi, Q.-Q.; Chi, S.-D.; Qu, S.; Luo, Y.-F.; He, Z.-M. SfgA renders Aspergillus flavus more stable to the external environment. J. Fungi 2022, 8, 638. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Zhang, F.; Nie, X.; Wang, X.; Yuan, J.; Zhuang, Z.; Wang, S. Essential APSES transcription factors for mycotoxin synthesis, fungal development, and pathogenicity in Aspergillus flavus. Front. Microbiol. 2017, 8, 2277. [Google Scholar] [CrossRef] [PubMed]
- Eom, T.-J.; Moon, H.; Yu, J.-H.; Park, H.-S. Characterization of the velvet regulators in Aspergillus flavus. J. Microbiol. 2018, 56, 893–901. [Google Scholar] [CrossRef]
- Wu, M.-Y.; Mead, M.E.; Kim, S.-C.; Rokas, A.; Yu, J.-H. WetA bridges cellular and chemical development in Aspergillus flavus. PLoS ONE 2017, 12, e0179571. [Google Scholar] [CrossRef]
- Adams, T.H.; Boylan, M.T.; Timberlake, W.E. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 1988, 54, 353–362. [Google Scholar] [CrossRef]
- Krijgsheld, P.; Bleichrodt, R.; van Veluw, G.; Wang, F.; Müller, W.; Dijksterhuis, J.; Wösten, H. Development in Aspergillus. Stud. Mycol. 2013, 74, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Boylan, M.T.; Mirabito, P.M.; Willett, C.E.; Zimmerman, C.R.; Timberlake, W.E. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol. Cell. Biol. 1987, 7, 3113–3118. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.-S.; Kim, Y.H.; Yu, J.-H. Proteomic analyses reveal the key roles of BrlA and AbaA in biogenesis of gliotoxin in Aspergillus fumigatus. Biochem. Biophys. Res. Commun. 2015, 463, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Yu, J.-H. AbaA and WetA govern distinct stages of Aspergillus fumigatus development. Microbiology 2011, 157, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-H.; Mah, J.-H.; Seo, J.-A. Growth and developmental control in the model and pathogenic Aspergilli. Eukaryot. Cell 2006, 5, 1577–1584. [Google Scholar] [CrossRef]
- Wu, M.-Y.; Mead, M.E.; Lee, M.-K.; Neuhaus, G.F.; Adpressa, D.A.; Martien, J.I.; Son, Y.-E.; Moon, H.; Amador-Noguez, D.; Han, K.-H.; et al. Transcriptomic, protein-DNA interaction, and metabolomic studies of VosA, VelB, and WetA in Aspergillus nidulans asexual spores. mBio 2021, 12, e03128-20. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-Y.; Mead, M.; Lee, M.-K.; Loss, E.M.O.; Kim, S.-C.; Rokas, A.; Yu, J.-H. Systematic dissection of the evolutionarily conserved WetA developmental regulator across a genus of filamentous fungi. mBio 2018, 9, e01130-18. [Google Scholar] [CrossRef] [PubMed]
- Clutterbuck, A.J. A Mutational analysis of conidial development in Aspergillus nidulans. Genetics 1969, 63, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Tokuoka, M.; Jin, F.J.; Takahashi, T.; Koyama, Y. Genetic analysis of conidiation regulatory pathways in koji-mold Aspergillus oryzae. Fungal Genet. Biol. 2010, 47, 10–18. [Google Scholar] [CrossRef]
- Yamada, O.; Lee, B.R.; Gomi, K.; Iimura, Y. Cloning and functional analysis of the Aspergillus oryzae conidiation regulator gene brlA by its disruption and misscheduled expression. J. Biosci. Bioeng. 1999, 87, 424–429. [Google Scholar] [CrossRef]
- Prade, R.; Timberlake, W. The Aspergillus nidulans brlA regulatory locus consists of overlapping transcription units that are individually required for conidiophore development. EMBO J. 1993, 12, 2439–2447. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Timberlake, W.E. Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics 1993, 133, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Etxebeste, O. Transcription factors in the fungus Aspergillus nidulans: Markers of genetic innovation, network rewiring and conflict between genomics and transcriptomics. J. Fungi 2021, 7, 600. [Google Scholar] [CrossRef]
- Twumasi-Boateng, K.; Yu, Y.; Chen, D.; Gravelat, F.N.; Nierman, W.C.; Sheppard, D.C. Transcriptional profiling identifies a role for BrlA in the response to nitrogen depletion and for StuA in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot. Cell 2009, 8, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Sewall, T.C.; Mims, C.W.; Timberlake, W.E. abaA controls phialide differentiation in Aspergillus nidulans. Plant Cell 1990, 2, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Andrianopoulos, A.; Timberlake, W.E. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 1994, 14, 2503–2515. [Google Scholar] [CrossRef] [PubMed]
- Aramayo, R.; Timberlake, W. The Aspergillus nidulans yA gene is regulated by abaA. EMBO J. 1993, 12, 2039–2048. [Google Scholar] [CrossRef]
- Mead, M.E.; Borowsky, A.T.; Joehnk, B.; Steenwyk, J.L.; Shen, X.-X.; Sil, A.; Rokas, A. Recurrent loss of abaA, a master regulator of asexual development in filamentous fungi, correlates with changes in genomic and morphological traits. Genome Biol. Evol. 2020, 12, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Marshall, M.A.; Timberlake, W.E. Aspergillus nidulans wetA activates spore-specific gene expression. Mol. Cell. Biol. 1991, 11, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Noble, L.M.; Andrianopoulos, A. Reproductive competence: A recurrent logic module in eukaryotic development. Proc. R. Soc. B Boil. Sci. 2013, 280, 20130819. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, D.; Gealt, M.; Pastushok, M. Gene control of developmental competence in Aspergillus nidulans. Dev. Biol. 1973, 34, 9–15. [Google Scholar] [CrossRef]
- Etxebeste, O.; Garzia, A.; Espeso, E.A.; Ugalde, U. Aspergillus nidulans asexual development: Making the most of cellular modules. Trends Microbiol. 2010, 18, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Wieser, J.; Lee, B.N.; Fondon, J.W.; Adams, T.H. Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr. Genet. 1994, 27, 62–69. [Google Scholar] [CrossRef]
- Lee, B.N.; Adams, T.H. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 1994, 8, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.N.; Adams, T.H. FluG and flbA function interdependently to initiate conidiophore development in Aspergillus nidulans through brlA beta activation. EMBO J. 1996, 15, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.I.; García, L.T.; Cortese, M.; Ugalde, U. The Early asexual development regulator fluG codes for a putative bifunctional enzyme. Front. Microbiol. 2019, 10, 778. [Google Scholar] [CrossRef]
- Lo, H.-C.; Entwistle, R.; Guo, C.-J.; Ahuja, M.; Szewczyk, E.; Hung, J.-H.; Chiang, Y.-M.; Oakley, B.R.; Wang, C.C.C. Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. J. Am. Chem. Soc. 2012, 134, 4709–4720. [Google Scholar] [CrossRef]
- Rodríguez-Urra, A.B.; Jiménez, C.; Nieto, M.I.; Rodríguez, J.; Hayashi, H.; Ugalde, U. Signaling the induction of sporulation involves the interaction of two secondary metabolites in Aspergillus nidulans. ACS Chem. Biol. 2012, 7, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.-K.; Scharfenstein, L.L.; Li, P.; Ehrlich, K.C. Aspergillus flavus VelB acts distinctly from VeA in conidiation and may coordinate with FluG to modulate sclerotial production. Fungal Genet. Biol. 2013, 58-59, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Na Lee, B.; Adams, T.H. Overexpression offIbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brIA and premature initiation of development. Mol. Microbiol. 1994, 14, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Kim, M.-J.; Yu, J.-H.; Shin, K.-S. Heterotrimeric G-protein signalers and RGSs in Aspergillus fumigatus. Pathogens 2020, 9, 902. [Google Scholar] [CrossRef]
- Etxebeste, O.; Ni, M.; Garzia, A.; Kwon, N.-J.; Fischer, R.; Yu, J.-H.; Espeso, E.A.; Ugalde, U. Basic-zipper-type transcription factor FlbB controls asexual development in Aspergillus nidulans. Eukaryot. Cell 2008, 7, 38–48. [Google Scholar] [CrossRef]
- Garzia, A.; Etxebeste, O.; Herrero-Garcia, E.; Fischer, R.; Espeso, E.A.; Ugalde, U. Aspergillus nidulansFlbE is an upstream developmental activator of conidiation functionally associated with the putative transcription factor FlbB. Mol. Microbiol. 2008, 71, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Arratia-Quijada, J.; Sánchez, O.; Scazzocchio, C.; Aguirre, J. FlbD, a Myb transcription factor of Aspergillus nidulans, is uniquely involved in both asexual and sexual differentiation. Eukaryot. Cell 2012, 11, 1132–1142. [Google Scholar] [CrossRef]
- Kwon, N.-J.; Garzia, A.; Espeso, E.A.; Ugalde, U.; Yu, J.-H. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 2010, 77, 1203–1219. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.; Shin, K.-S.; Wang, T.; Yu, J.-H. Aspergillus fumigatus flbB encodes two basic leucine zipper domain (bZIP) proteins required for proper asexual development and gliotoxin production. Eukaryot. Cell 2010, 9, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Otamendi, A.; Perez-De-Nanclares-Arregi, E.; Oiartzabal-Arano, E.; Cortese, M.S.; Espeso, E.A.; Etxebeste, O. Developmental regulators FlbE/D orchestrate the polarity site-to-nucleus dynamics of the fungal bZIP transcription factor FlbB. Experientia 2019, 76, 4369–4390. [Google Scholar] [CrossRef]
- Herrero-Garcia, E.; Perez-De-Nanclares-Arregi, E.; Cortese, M.; Markina-Iñarrairaegui, A.; Oiartzabal-Arano, E.; Etxebeste, O.; Ugalde, U.; Espeso, E.A. Tip-to-nucleus migration dynamics of the asexual development regulator FlbB in vegetative cells. Mol. Microbiol. 2015, 98, 607–624. [Google Scholar] [CrossRef] [PubMed]
- Bayram, Ö.; Braus, G.H. Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Yu, J.-H. Velvet regulators in Aspergillus spp. Microbiol. Biotechnol. Lett. 2016, 44, 409–419. [Google Scholar] [CrossRef]
- Bayram, O.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.-J.; Keller, N.P.; Yu, J.-H.; et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar] [CrossRef] [PubMed]
- Bayram, S.; Bayram, Ö.; Valerius, O.; Park, H.S.; Irniger, S.; Gerke, J.; Ni, M.; Han, K.-H.; Yu, J.-H.; Braus, G.H. LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 2010, 6, e1001226. [Google Scholar] [CrossRef]
- Kim, H.-S.; Han, K.-Y.; Kim, K.-J.; Han, D.-M.; Jahng, K.-Y.; Chae, K.-S. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 2002, 37, 72–80. [Google Scholar] [CrossRef]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 2004, 3, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.; Bok, J.; Chung, D.; Perrin, R.M.; Shwab, E.K. LaeA, a global regulator of Aspergillus toxins. Med Mycol. 2006, 44, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.-K.; Scharfenstein, L.L.; Ehrlich, K.C.; Wei, Q.; Bhatnagar, D.; Ingber, B.F. Effects of laeA deletion on Aspergillus flavus conidial development and hydrophobicity may contribute to loss of aflatoxin production. Fungal Biol. 2011, 116, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Lara-Rojas, F.; Sánchez, O.; Kawasaki, L.; Aguirre, J. Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol. Microbiol. 2011, 80, 436–454. [Google Scholar] [CrossRef] [PubMed]
- Leiter, É.; Emri, T.; Pákozdi, K.; Hornok, L.; Pócsi, I. The impact of bZIP Atf1ortholog global regulators in fungi. Appl. Microbiol. Biotechnol. 2021, 105, 5769–5783. [Google Scholar] [CrossRef]
- Garrido-Bazán, V.; Jaimes-Arroyo, R.; Sánchez, O.; Lara-Rojas, F.; Aguirre, J. SakA and MpkC stress MAPKs show opposite and common functions during stress responses and development in Aspergillus nidulans. Front. Microbiol. 2018, 9, 2518. [Google Scholar] [CrossRef]
- Bailey, C.; Arst, H.N. Carbon Catabolite Repression in Aspergillus nidulans. JBIC J. Biol. Inorg. Chem. 1975, 51, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Zheng, W.; Islam, W.; Arif, M.; Abubakar, Y.S.; Wang, Z.; Lu, G. Carbon Catabolite Repression in Filamentous Fungi. Int. J. Mol. Sci. 2017, 19, 48. [Google Scholar] [CrossRef]
- Ries, L.N.; Beattie, S.R.; Espeso, E.A.; Cramer, R.A.; Goldman, G.H. Diverse Regulation of the CreA Carbon Catabolite Repressor in Aspergillus nidulans. Genetics 2016, 203, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Lee, S.C.; Cardenas, M.E.; Heitman, J. Calcium-calmodulin-calcineurin signaling: A globally conserved virulence cascade in eukaryotic microbial pathogens. Cell Host Microbe 2019, 26, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Cramer, R.A.; Perfect, B.Z.; Pinchai, N.; Park, S.; Perlin, D.S.; Asfaw, Y.G.; Heitman, J.; Perfect, J.R.; Steinbach, W.J. Calcineurin target CrzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryot. Cell 2008, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Juvvadi, P.R.; Lamoth, F.; Steinbach, W.J. Calcineurin as a multifunctional regulator: Unraveling novel functions in fungal stress responses, hyphal growth, drug resistance, and pathogenesis. Fungal Biol. Rev. 2014, 28, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Spielvogel, A.; Findon, H.; Arst, H.N.; Araújo-Bazán, L.; Hernández-Ortíz, P.; Stahl, U.; Meyer, V.; Espeso, E.A. Two zinc finger transcription factors, CrzA and SltA, are involved in cation homoeostasis and detoxification in Aspergillus nidulans. Biochem. J. 2008, 414, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ortiz, P.; Espeso, E.A. Spatiotemporal dynamics of the calcineurin target CrzA. Cell. Signal. 2017, 29, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, W.J.; Cramer, R.A.; Perfect, B.Z.; Asfaw, Y.G.; Sauer, T.C.; Najvar, L.K.; Kirkpatrick, W.R.; Patterson, T.F.; Benjamin, D.K.; Heitman, J.; et al. Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 2006, 5, 1091–1103. [Google Scholar] [CrossRef] [Green Version]
- De Nadal, E.; Posas, F. The HOG pathway and the regulation of osmoadaptive responses in yeast. FEMS Yeast Res. 2022, 22, foac013. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Li, R. Current Understanding of HOG-MAPK pathway in Aspergillus fumigatus. Mycopathologia 2012, 175, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.A.; Morgan, B.A.; Quinn, J. Stress signalling to fungal stress-activated protein kinase pathways. FEMS Microbiol. Lett. 2010, 306, 1–8. [Google Scholar] [CrossRef]
- Emri, T.; Szarvas, V.; Orosz, E.; Antal, K.; Park, H.; Han, K.-H.; Yu, J.-H.; Pócsi, I. Core oxidative stress response in Aspergillus nidulans. BMC Genom. 2015, 16, 478. [Google Scholar] [CrossRef]
- Ramamoorthy, V.; Dhingra, S.; Kincaid, A.; Shantappa, S.; Feng, X.; Calvo, A.M. The putative C2H2 transcription factor MtfA is a novel regulator of secondary metabolism and morphogenesis in Aspergillus nidulans. PLoS ONE 2013, 8, e74122. [Google Scholar] [CrossRef]
- Smith, T.D.; Calvo, A.M. The mtfA transcription factor gene controls morphogenesis, gliotoxin production, and virulence in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot. Cell 2014, 13, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-R.; Chae, K.-S.; Han, K.-H.; Han, D.-M. The nsdC Gene Encoding a Putative C2H2-Type Transcription Factor Is a Key Activator of Sexual Development in Aspergillus nidulans. Genetics 2009, 182, 771–783. [Google Scholar] [CrossRef]
- Han, K.; Yu, J.; Chae, K.; Jahng, K. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol. Microbiol. 2001, 41, 299–309. [Google Scholar] [CrossRef]
- Lee, M.-K.; Kwon, N.-J.; Lee, I.-S.; Jung, S.; Kim, S.-C.; Yu, J.-H. Negative regulation and developmental competence in Aspergillus. Sci. Rep. 2016, 6, 28874. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.K.; Mack, B.M.; Wei, Q.; Bland, J.; Bhatnagar, D.; Cary, J.W. RNA sequencing of an nsdC mutant reveals global regulation of secondary metabolic gene clusters in Aspergillus flavus. Microbiol. Res. 2016, 182, 150–161. [Google Scholar] [CrossRef]
- Seo, J.-A.; Guan, Y.; Yu, J.-H. Suppressor Mutations Bypass the Requirement of fluG for Asexual Sporulation and Sterigmatocystin Production in Aspergillus nidulans. Genetics 2003, 165, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-A.; Guan, Y.; Yu, J.-H. FluG-Dependent Asexual Development in Aspergillus nidulans Occurs via Derepression. Genetics 2006, 172, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.Y.; Toennis, T.M.; Adams, T.H.; Miller, B.L. Isolation and transcriptional characterization of a morphological modifier: The Aspergillus nidulans stunted (stuA) gene. Mol. Gen. Genet. MGG 1991, 227, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.Y.; Wu, J.; Miller, B.L. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 1992, 6, 1770–1782. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, D.C.; Doedt, T.; Chiang, L.Y.; Kim, H.; Chen, D.; Nierman, W.C.; Filler, S.G. The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol. Biol. Cell 2005, 16, 5866–5879. [Google Scholar] [CrossRef]
- Wang, P.; Xu, J.; Chang, P.-K.; Liu, Z.; Kong, Q. New Insights of transcriptional regulator AflR in Aspergillus flavus physiology. Microbiol. Spectr. 2022, 10, e00791-21. [Google Scholar] [CrossRef]
- Han, X.; Qiu, M.; Wang, B.; Yin, W.-B.; Nie, X.; Qin, Q.; Ren, S.; Yang, K.; Zhang, F.; Zhuang, Z.; et al. Functional analysis of the nitrogen metabolite repression regulator gene nmrA in Aspergillus flavus. Front. Microbiol. 2016, 7, 1794. [Google Scholar] [CrossRef] [PubMed]
- Mengjuan, Z.; Guanglan, L.; Xiaohua, P.; Weitao, S.; Can, T.; Xuan, C.; Yanling, Y.; Zhenhong, Z. The PHD transcription factor Cti6 is involved in the fungal colonization and aflatoxin B1 biological synthesis of Aspergillus flavus. IMA Fungus 2021, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Cary, J.W.; Harris-Coward, P.; Scharfenstein, L.; Mack, B.M.; Chang, P.-K.; Wei, Q.; Lebar, M.; Carter-Wientjes, C.; Majumdar, R.; Mitra, C.; et al. The Aspergillus flavus homeobox gene, hbx1, is required for development and aflatoxin production. Toxins 2017, 9, 315. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Wiemann, P.; Garvey, G.S.; Lim, F.Y.; Haas, B.; Wortman, J.; Keller, N.P. Illumina identification of RsrA, a conserved C2H2 transcription factor coordinating the NapA mediated oxidative stress signaling pathway in Aspergillus. BMC Genom. 2014, 15, 1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Yang, G.; Zhang, D.; Liu, Y.; Li, Y.; Lin, G.; Guo, Z.; Wang, S.; Zhuang, Z. The PHD transcription factor Rum1 regulates morphogenesis and aflatoxin biosynthesis in Aspergillus flavus. Toxins 2018, 10, 301. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Xu, G.; Geng, L.; Lu, X.; Yang, K.; Yuan, J.; Nie, X.; Zhuang, Z.; Wang, S. The stress response regulator AflSkn7 influences morphological development, stress response, and pathogenicity in the fungus Aspergillus flavus. Toxins 2016, 8, 202. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.-E.; Cho, H.-J.; Lee, M.-K.; Park, H.-S. Characterizing the role of Zn cluster family transcription factor ZcfA in governing development in two Aspergillus species. PLoS ONE 2020, 15, e0228643. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Genes | Domain(s) | Description | Ref. |
|---|---|---|---|
| AbaA | TEA/ATTS | Regulator for sterigmata formation | - |
| AtfA | bZIP | Developmental activator | [31] |
| AtfB | bZIP | Developmental activator | [31] |
| BrlA | C2H2 zinc finger | Initiator for conidiogenesis | - |
| CreA | C2H2 zinc finger | Developmental activator | [32] |
| CrzA | C2H2 zinc finger | Developmental activator | [33] |
| FlbA | RGS | Upstream developmental activator | [34] |
| FlbB | Basic leucine zipper | Upstream developmental activator | - |
| FlbC | C2H2 zinc finger | Upstream dev. activator | - |
| FlbD | Myb-like DNA binding | Upstream dev. activator | - |
| FlbE | - | Upstream dev. activator | - |
| FluG | Amidohydrolase and GS | Upstream dev. activator | [35] |
| HogA | Protein kinase | Modulator for conidiation | [36] |
| LaeA | SAM | Developmental repressor | [37] |
| MsnA | C2H2 zinc finger | Developmental repressor | [38] |
| MtfA | C2H2 zinc finger | Developmental repressor | [39] |
| NsdC | C2H2 zinc finger | Developmental repressor | [40] |
| NsdD | GATA-type zinc-finger | Developmental repressor | [40] |
| SfgA | Zn2Cys6 | Developmental repressor | [41] |
| StuA | APSES | Modulator for conidiophore formation | [42] |
| VeA | Velvet | Developmental activator | [37] |
| VelB | Velvet | Developmental activator Regulator for conidial maturation | [43] |
| VelC | Velvet | - | [43] |
| VelD | Velvet | - | [43] |
| VosA | Velvet | Regulator for conidial maturation | [43] |
| WetA | ESC1/WetA-related | Regulator for conidial maturation | [44] |
| Genes | Asexual Development | Sexual Structure Formation | Ref. | ||
|---|---|---|---|---|---|
| Conidia Formation | brlA Expression | ||||
| FlbB | ΔAniflbB | decrease | absence | not determined | [77] |
| ΔAflflbB | decrease | decrease | decrease | - | |
| FlbC | ΔAniflbC | decrease | delay | increase | [80] |
| ΔAflflbC | decrease | decrease | decrease | - | |
| FlbD | ΔAniflbD | decrease | delay | decrease | [79] |
| ΔAflflbD | decrease | decrease | decrease | - | |
| FlbE | ΔAniflbE | decrease | delay | not determined | [78] |
| ΔAflflbE | decrease | decrease | decrease | - | |
| Genes | Domain(s) | Phenotype of Deletion Mutant | Ref. |
|---|---|---|---|
| AflR | Zn2Cys6 domain | Decrease conidiophore production | [119] |
| AreA | GATA zinc finger domain | Decrease conidiophore production and brlA expression | [120] |
| Cti6 | PHD domain | Decrease conidiophore production and brlA expression | [121] |
| Hbx1 | Homeodomain | Loss of conidiophore, decrease brlA expression | [122] |
| RafA | APSES | Decrease conidiophore production, increase brlA expression | [42] |
| RsrA | C2H2 zinc finger | Decrease conidiophore production | [123] |
| Rum1 | PHD domain | Increase conidiophore production and brlA expression | [124] |
| Skn7 | Heat-shock transcription factor-like DNA-binding domain | Abnormal conidiophore, decrease conidiophore production | [125] |
| ZcfA | Zn2Cys6 domain | Increase conidiophore production | [126] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, H.-J.; Son, S.-H.; Chen, W.; Son, Y.-E.; Lee, I.; Yu, J.-H.; Park, H.-S. Regulation of Conidiogenesis in Aspergillus flavus. Cells 2022, 11, 2796. https://doi.org/10.3390/cells11182796
Cho H-J, Son S-H, Chen W, Son Y-E, Lee I, Yu J-H, Park H-S. Regulation of Conidiogenesis in Aspergillus flavus. Cells. 2022; 11(18):2796. https://doi.org/10.3390/cells11182796
Chicago/Turabian StyleCho, He-Jin, Sung-Hun Son, Wanping Chen, Ye-Eun Son, Inhyung Lee, Jae-Hyuk Yu, and Hee-Soo Park. 2022. "Regulation of Conidiogenesis in Aspergillus flavus" Cells 11, no. 18: 2796. https://doi.org/10.3390/cells11182796
APA StyleCho, H.-J., Son, S.-H., Chen, W., Son, Y.-E., Lee, I., Yu, J.-H., & Park, H.-S. (2022). Regulation of Conidiogenesis in Aspergillus flavus. Cells, 11(18), 2796. https://doi.org/10.3390/cells11182796