Mechanistic Understanding of the Olfactory Neuroepithelium Involvement Leading to Short-Term Anosmia in COVID-19 Using the Adverse Outcome Pathway Framework
Abstract
:1. Introduction
1.1. Anosmia and COVID-19
1.2. The Olfactory Mucosa
1.2.1. Anatomy
1.2.2. Olfactory Physiology
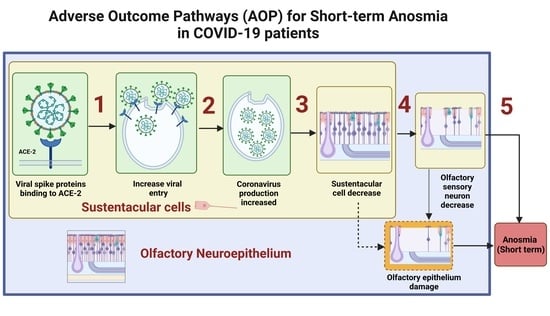
2. Underlying Biological Mechanisms Leading to Short-Term Anosmia in COVID-19
3. OE Damage Underlying Short-Term Anosmia
3.1. S Protein Binding to ACE2 in SUS Cells Leads to Increased Viral Entry
3.1.1. Biological Plausibility
3.1.2. Empirical Evidence
3.1.3. Modulating Factors
3.1.4. Overall Assessment
| KE1739 and KE1738 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Observations | Model Organism | Type of Study | Ref. | |||||
| Hu | Mo | Ha | S1 | S2 | S3 | S4 | ||
| ACE2 is expressed in the SUS cells of the OE to elucidate the symptoms of SARS-CoV-2-induced anosmia via the olfactory pathway | ☒ | ☐ | ☐ | ☒ | ☐ | ☐ | ☐ | [49] |
| Subsets of human OE SUS cells (basal and Bowman’s gland cells) express the SARS-CoV-2 receptor ACE2 | ☒ | ☐ | ☐ | ☒ | ☐ | ☐ | ☐ | [53] |
| ACE2 expression confirmedin OE inhumans | ☒ | ☐ | ☐ | ☒ | ☐ | ☐ | ☐ | [63] |
| The upper airway is the initial site of SARS-CoV-2 infection based on data on ACE2 expression in the SUS cells | ☒ | ☐ | ☐ | ☒ | ☐ | ☐ | ☐ | [51] |
| ACE2 protein is highly expressed in a subset of SUS cells in human olfactory tissues | ☒ | ☐ | ☐ | ☒ | ☐ | ☐ | ☐ | [52] |
| Meta-analysis indicates that ACE2is enriched within OE subsets | ☒ | ☐ | ☐ | ☐ | ☐ | ☐ | ☒ | [64] |
| Single-cell RNA-sequencing uncovers putative targets of SARS-CoV-2 among tissue-resident cell subsets. | ☐ | ☒ | ☐ | ☐ | ☐ | ☐ | ☒ | [64] |
| Subsets of mouse OE SUS cells,horizontal basal cells (HBCs), and Bowman’s gland cells express the ACE2 receptor | ☐ | ☒ | ☐ | ☒ | ☐ | ☐ | ☐ | [53] |
| ACE2 is expressed in thesupporting cells and Bowman’s gland of the mouse Olfactory Mucosa (OM), except for the cilia in the respiratory epithelium of mice | ☐ | ☒ | ☐ | ☒ | ☐ | ☐ | ☐ | [63] |
| ACE2 protein is highly expressed in a subset of SUS cells in human and mouse olfactory tissues | ☐ | ☒ | ☐ | ☒ | ☐ | ☐ | ☐ | [52] |
| SUS cells are identified as the main target cells of SARS-CoV-2 in the OE, which highly express ACE2 | ☒ | ☐ | ☐ | ☐ | ☐ | ☒ | ☐ | [41] |
3.2. Viral Entry in SUS Cells Leads to Increased SARS-CoV-2 Production
3.2.1. Biological Plausibility
3.2.2. Empirical Evidence
3.2.3. Modulating Factors
3.2.4. Overall Assessment
3.3. SARS-CoV-2 Production Leads to SUS Cells Decrease
3.3.1. Biological Plausibility
3.3.2. Empirical Evidence
3.3.3. Modulating Factors
3.3.4. Overall Assessment
3.4. SUS Cells Decrease Leads to Reduction in the Olfactory Sensory Neurons
3.4.1. Biological Plausibility
3.4.2. Empirical Evidence
| 12 hpi | 1 dpi | 2 dpi | 3 dpi | 4 dpi | 5 dpi | 7 dpi | 10 dpi | 14 dpi | 21 dpi | Olfactory Sensory Neurons Downregulation and Decrease (KE Upstream) | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ☐ | ☒ | ☒ | ☐ | ☒ | ☐ | ☐ | ☐ | ☐ | ☐ | Delayed downregulation of OSN-specific markers and their precursors is observed by 4 dpi | [74] |
| ☐ | ☐ | ☐ | ☐ | ☒ | ☐ | ☐ | ☐ | ☐ | ☐ | SARS-CoV-2 nuclear protein colocalizes within olfactory sensory neuron marker OMP is detected at 4 dpi | [80] |
| ☐ | ☐ | ☒ | ☐ | ☐ | ☐ | ☐ | ☐ | ☐ | ☐ | OSNs cilia are severely impaired after infection at 2 dpi. | [50] |
| ☐ | ☐ | ☐ | ☐ | ☒ | ☐ | ☐ | ☐ | ☐ | ☐ | Infected OMP+ mature OSNs were discovered at 4 dpi. | [65] |
| 12 hpi | 1 dpi | 2 dpi | 3 dpi | 4 dpi | 5 dpi | 7 dpi | 10 dpi | 14 dpi | 21 dpi | OE Damage (KE Downstream) | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ☒ | ☐ | ☒ | ☐ | ☒ | ☐ | ☒ | ☐ | ☐ | ☐ | SARS-CoV-2 N protein (NP) is scattered to indicate epithelial cell damage upon infection; damage occurs 12 h postinfection and onwards. | [80] |
| ☐ | ☒ | ☒ | ☐ | ☒ | ☐ | ☐ | ☐ | ☐ | ☐ | OE tissue damage but nonsignificant increase in apoptotic markers at 4 dpi. | [74] |
| ☐ | ☐ | ☒ | ☐ | ☒ | ☐ | ☒ | ☒ | ☒ | ☐ | SARS-CoV-2 strains UCN1 and UCN19 induce massive damage to the OE at 2 dpi, but it becomes partially healed at 14 dpi. | [50] |
| ☐ | ☐ | ☒ | ☐ | ☒ | ☐ | ☐ | ☐ | ☒ | ☐ | The OE shows loss of ciliation as early as 2 dpi. | [65] |
| ☐ | ☐ | ☐ | ☒ | ☐ | ☒ | ☐ | ☒ | ☐ | ☒ | Prominent nasal discharge from the OE was observed as early as 3 dpi but the nasal cavity normalized at the time points of 10 and 21 dpi | [81] |
3.4.3. Modulating Factors
3.5. Olfactory Sensory Neuron Genes Downregulation Leads to Short-Term Anosmia
3.5.1. Biological Plausibility
3.5.2. Empirical Evidence
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Disclaimer
References
- OECD. Guidance Document for the Use of Adverse Outcome Pathways in Developing Integrated Approaches to Testing and Assessment (IATA); OECD: Paris, France, 2017. [Google Scholar] [CrossRef]
- Tollefsen, K.E.; Scholz, S.; Cronin, M.T.; Edwards, S.W.; de Knecht, J.; Crofton, K.; Garcia-Reyero, N.; Hartung, T.; Worth, A.; Patlewicz, G. Applying Adverse Outcome Pathways (AOPs) to Support Integrated Approaches to Testing and Assessment (IATA). Regul. Toxicol. Pharmacol. 2014, 70, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Tsamou, M.; Pistollato, F.; Roggen, E.L. A Tau-Driven Adverse Outcome Pathway Blueprint Toward Memory Loss in Sporadic (Late-Onset) Alzheimer’s Disease with Plausible Molecular Initiating Event Plug-Ins for Environmental Neurotoxicants. J. Alzheimers Dis. 2021, 81, 459–485. [Google Scholar] [CrossRef] [PubMed]
- Nymark, P.; Sachana, M.; Leite, S.B.; Sund, J.; Krebs, C.E.; Sullivan, K.; Edwards, S.; Viviani, L.; Willett, C.; Landesmann, B.; et al. Systematic Organization of COVID-19 Data Supported by the Adverse Outcome Pathway Framework. Front. Public Health 2021, 9, 638605. [Google Scholar] [CrossRef] [PubMed]
- AOP-Wiki. Available online: https://aopwiki.org/ (accessed on 14 June 2022).
- Becker, R.A.; Ankley, G.T.; Edwards, S.W.; Kennedy, S.W.; Linkov, I.; Meek, B.; Sachana, M.; Segner, H.; van der Burg, B.; Villeneuve, D.L.; et al. Increasing Scientific Confidence in Adverse Outcome Pathways: Application of Tailored Bradford-Hill Considerations for Evaluating Weight of Evidence. Regul. Toxicol. Pharmacol. 2015, 72, 514–537. [Google Scholar] [CrossRef] [PubMed]
- Hummel, T.; Whitcroft, K.L.; Andrews, P.; Altundag, A.; Cinghi, C.; Costanzo, R.M.; Damm, M.; Frasnelli, J.; Gudziol, H.; Gupta, N.; et al. Position Paper on Olfactory Dysfunction. Rhinology 2017, 54, 1–30. [Google Scholar] [CrossRef]
- Vargas-Gandica, J.; Winter, D.; Schnippe, R.; Rodriguez-Morales, A.G.; Mondragon, J.; Antezana, J.P.E.; Trelles-Thorne, M.D.P.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J.; Paniz-Mondolfi, A. Ageusia and Anosmia, a Common Sign of COVID-19? A Case Series from Four Countries. J. NeuroVirol. 2020, 26, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Mutiawati, E.; Fahriani, M.; Mamada, S.S.; Fajar, J.K.; Frediansyah, A.; Maliga, H.A.; Ilmawan, M.; Bin Emran, T.; Ophinni, Y.; Ichsan, I.; et al. Anosmia and Dysgeusia in SARS-CoV-2 Infection: Incidence and Effects on COVID-19 Severity and Mortality, and the Possible Pathobiology Mechanisms-a Systematic Review and Meta-Analysis. F1000Research 2021, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Butowt, R.; von Bartheld, C.S. Anosmia in COVID-19: Underlying Mechanisms and Assessment of an Olfactory Route to Brain Infection. Neuroscientist 2021, 27, 582–603. [Google Scholar] [CrossRef] [PubMed]
- von Bartheld, C.S.; Hagen, M.M.; Butowt, R. Prevalence of Chemosensory Dysfunction in COVID-19 Patients: A Systematic Review and Meta-Analysis Reveals Significant Ethnic Differences. medRxiv 2020, 11, 2944–2961. [Google Scholar] [CrossRef] [PubMed]
- Niemi, M.E.K.; Karjalainen, J.; Liao, R.G.; Neale, B.M.; Daly, M.; Ganna, A.; Pathak, G.A.; Andrews, S.J.; Kanai, M.; Veerapen, K.; et al. Mapping the Human Genetic Architecture of COVID-19. Nature 2021, 600, 472–477. [Google Scholar] [CrossRef]
- von Bartheld, C.S.; Hagen, M.M.; Butowt, R. The D614G Virus Mutation Enhances Anosmia in COVID-19 Patients: Evidence from a Systematic Review and Meta-Analysis of Studies from South Asia. ACS Chem. Neurosci. 2021, 12, 3535–3549. [Google Scholar] [CrossRef] [PubMed]
- See, A.; Ko, K.K.K.; Toh, S.T. Epidemiological Analysis in Support of Hypothesis That D614G Virus Mutation Is a Major Contributing Factor to Chemosensory Dysfunction in COVID-19 Patients. Eur. Arch. Oto-Rhino-Laryngol. 2021, 278, 3595–3596. [Google Scholar] [CrossRef] [PubMed]
- Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants/ (accessed on 8 September 2022).
- Maisa, A.; Spaccaferri, G.; Fournier, L.; Schaeffer, J.; Deniau, J.; Rolland, P.; Coignard, B.; Andrieu, A.; Broustal, O.; Chene, S.; et al. First Cases of Omicron in France Are Exhibiting Mild Symptoms, November 2021–January 2022. Infect. Dis. Now 2022, 52, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Boscolo-Rizzo, P.; Tirelli, G.; Meloni, P.; Hopkins, C.; Madeddu, G.; de Vito, A.; Gardenal, N.; Valentinotti, R.; Tofanelli, M.; Borsetto, D.; et al. Coronavirus Disease 2019 (COVID-19)–Related Smell and Taste Impairment with Widespread Diffusion of Severe Acute Respiratory Syndrome–Coronavirus-2 (SARS-CoV-2) Omicron Variant. Int. Forum. Allergy Rhinol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Pouwels, K.B.; Pritchard, E.; Matthews, P.C.; Stoesser, N.; Eyre, D.W.; Vihta, K.D.; House, T.; Hay, J.; Bell, J.I.; Newton, J.N.; et al. Effect of Delta Variant on Viral Burden and Vaccine Effectiveness against New SARS-CoV-2 Infections in the UK. Nat. Med. 2021, 27, 2127–2135. [Google Scholar] [CrossRef] [PubMed]
- Klimek, L.; Hagemann, J.; Hummel, T.; Altundag, A.; Hintschich, C.; Stielow, S.; Bousquet, J. Olfactory Dysfunction Is More Severe in Wild-Type SARS-CoV-2 Infection than in the Delta Variant (B.1.617.2). World Allergy Organ. J. 2022, 15, 100653. [Google Scholar] [CrossRef] [PubMed]
- Coronavirus (COVID-19) Infection Survey: Characteristics of People Testing Positive for COVID-19 in England, 27 January 2021-Office for National Statistics. Available online: https://www.ons.gov.uk/releases/coronaviruscovid19infectionsurveycharacteristicsofpeopletestingpositiveforcovid19inengland27january2021 (accessed on 13 June 2022).
- Vihta1, K.D.; Pouwels, K.B.; Peto1, T.E.; Pritchard, E.; House, T.; Studley, R.; Rourke, E.; Cook, D.; Diamond, I.; Crook1, D.; et al. Omicron-Associated Changes in SARS-CoV-2 Symptoms in the United Kingdom. Clin. Infect. Dis. 2022, ciac613. [Google Scholar] [CrossRef] [PubMed]
- Butowt, R.; Bilińska, K.; von Bartheld, C. Why Does the Omicron Variant Largely Spare Olfactory Function? Implications for the Pathogenesis of Anosmia in Coronavirus Disease 2019. J. Infect. Dis. 2022, jiac113. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Sevilla, J.J.; Güerri-Fernádez, R.; Bertran Recasens, B. Is There Less Alteration of Smell Sensation in Patients With Omicron SARS-CoV-2 Variant Infection? Front. Med. 2022, 9, 1044. [Google Scholar] [CrossRef] [PubMed]
- Clerbaux, L.-A.; Albertini, M.C.; Amigó, N.; Beronius, A.; Bezemer, G.F.G.; Coecke, S.; Daskalopoulos, E.P.; del Giudice, G.; Greco, D.; Grenga, L.; et al. Factors Modulating COVID-19: A Mechanistic Understanding Based on the Adverse Outcome Pathway Framework. Preprints 2022, 11, 4464. [Google Scholar] [CrossRef]
- Lee, Y.; Min, P.; Lee, S.; Kim, S.W. Prevalence and Duration of Acute Loss of Smell or Taste in COVID-19 Patients. J. Korean Med. Sci. 2020, 35, e174. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.K.J.; Han, R.; Zhao, J.J.; Tan, N.K.W.; Quah, E.S.H.; Tan, C.J.-W.; Chan, Y.H.; Teo, N.W.Y.; Charn, T.C.; See, A.; et al. Prognosis and Persistence of Smell and Taste Dysfunction in Patients with Covid-19: Meta-Analysis with Parametric Cure Modelling of Recovery Curves. BMJ 2022, 378, e069503. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, R.; Dini, M.; Rosci, C.; Capozza, A.; Groppo, E.; Reitano, M.R.; Allocco, E.; Poletti, B.; Brugnera, A.; Bai, F.; et al. One-Year Cognitive Follow-up of COVID-19 Hospitalized Patients. Eur. J. Neurol. 2022, 29, 2006–2014. [Google Scholar] [CrossRef] [PubMed]
- Sanli, D.E.T.; Altundag, A.; Ylldlrlm, D.; Kandemirli, S.G.; Sanli, A.N. Comparison of Olfactory Cleft Width and Volumes in Patients with COVID-19 Anosmia and COVID-19 Cases Without Anosmia. ORL 2022, 84, 1–9. [Google Scholar] [CrossRef] [PubMed]
- da Costa, K.V.; Carnaúba, A.T.L.; Rocha, K.W.; de Andrade, K.C.L.; Ferreira, S.M.S.; Menezes, P.D.L. Olfactory and Taste Disorders in COVID-19: A Systematic Review. Braz. J. Otorhinolaryngol. 2020, 86, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Van Riel, D.; Verdijk, R.; Kuiken, T. The Olfactory Nerve: A Shortcut for Influenza and Other Viral Diseases into the Central Nervous System. J. Pathol. 2015, 235, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Salazar, I.; Sanchez-Quinteiro, P.; Barrios, A.W.; López Amado, M.; Vega, J.A. Anatomy of the Olfactory Mucosa. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 164, pp. 47–65. [Google Scholar]
- Pinto, J.M. Olfaction. Proc. Am. Thorac. Soc. 2011, 8, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Rawson, N.E. Olfactory Loss in Aging. Sci. Aging Knowl. Environ. 2006, 2006, pe6. [Google Scholar] [CrossRef]
- Ma, M. Encoding Olfactory Signals via Multiple Chemosensory Systems. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 463–480. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, S.; Goldstein, B.J. Pathophysiology of Olfactory Disorders and Potential Treatment Strategies. Curr. Otorhinolaryngol. Rep. 2016, 4, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Han, A.Y.; Mukdad, L.; Long, J.L.; Lopez, I.A. Anosmia in COVID-19: Mechanisms and Significance. Chem. Senses 2020, 45, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Cui, C.; Hautefort, C.; Haehner, A.; Zhao, J.; Yao, Q.; Zeng, H.; Nisenbaum, E.J.; Liu, L.; Zhao, Y.; et al. Olfactory and Gustatory Dysfunction as an Early Identifier of COVID-19 in Adults and Children: An International Multicenter Study. Otolaryngol.-Head Neck Surg. 2020, 163, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Gane, S.B.C.; Kelly, C.; Hopkins, C. Isolated Sudden Onset Anosmia in COVID-19 Infection. A Novel Syndrome? Rhinology 2020, 58, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Eliezer, M.; Hautefort, C.; Hamel, A.L.; Verillaud, B.; Herman, P.; Houdart, E.; Eloit, C. Sudden and Complete Olfactory Loss of Function as a Possible Symptom of COVID-19. JAMA Otolaryngol. Head Neck Surg. 2020, 146, 674–675. [Google Scholar] [CrossRef]
- Jalessi, M.; Barati, M.; Rohani, M.; Amini, E.; Ourang, A.; Azad, Z.; Hosseinzadeh, F.; Cavallieri, F.; Ghadirpour, R.; Valzania, F.; et al. Frequency and Outcome of Olfactory Impairment and Sinonasal Involvement in Hospitalized Patients with COVID-19. Neurol. Sci. 2020, 41, 2331–2338. [Google Scholar] [CrossRef]
- Khan, M.; Yoo, S.J.; Clijsters, M.; Backaert, W.; Vanstapel, A.; Speleman, K.; Lietaer, C.; Choi, S.; Hether, T.D.; Marcelis, L.; et al. Visualizing in Deceased COVID-19 Patients How SARS-CoV-2 Attacks the Respiratory and Olfactory Mucosae but Spares the Olfactory Bulb. Cell 2021, 184, 5932–5949.e15. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nagata, N.; Shirato, K.; Kawase, M.; Takeda, M.; Taguchi, F. Efficient Activation of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein by the Transmembrane Protease TMPRSS2. J. Virol. 2010, 84, 12658–12664. [Google Scholar] [CrossRef]
- Shulla, A.; Heald-Sargent, T.; Subramanya, G.; Zhao, J.; Perlman, S.; Gallagher, T. A Transmembrane Serine Protease Is Linked to the Severe Acute Respiratory Syndrome Coronavirus Receptor and Activates Virus Entry. J. Virol. 2011, 85, 873–882. [Google Scholar] [CrossRef]
- Glowacka, I.; Bertram, S.; Müller, M.A.; Allen, P.; Soilleux, E.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence That TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J. Virol. 2011, 85, 4122–4134. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Hofmann, H.; Pyrc, K.; van der Hoek, L.; Geier, M.; Berkhout, B.; Pöhlmann, S. Human Coronavirus NL63 Employs the Severe Acute Respiratory Syndrome Coronavirus Receptor for Cellular Entry. Proc. Natl. Acad. Sci. USA 2005, 102, 7988–7993. [Google Scholar] [CrossRef] [PubMed]
- Simmons, G.; Gosalia, D.N.; Rennekamp, A.J.; Reeves, J.D.; Diamond, S.L.; Bates, P. Inhibitors of Cathepsin L Prevent Severe Acute Respiratory Syndrome Coronavirus Entry. Proc. Natl. Acad. Sci. USA 2005, 102, 11876–11881. [Google Scholar] [CrossRef] [PubMed]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The Spike Glycoprotein of the New Coronavirus 2019-NCoV Contains a Furin-like Cleavage Site Absent in CoV of the Same Clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef] [PubMed]
- Klingenstein, M.; Klingenstein, S.; Neckel, P.H.; Mack, A.F.; Wagner, A.P.; Kleger, A.; Liebau, S.; Milazzo, A. Evidence of SARS-CoV2 Entry Protein ACE2 in the Human Nose and Olfactory Bulb. Cells Tissues Organs 2020, 209, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Bryche, B.; St Albin, A.; Murri, S.; Lacôte, S.; Pulido, C.; Ar Gouilh, M.; Lesellier, S.; Servat, A.; Wasniewski, M.; Picard-Meyer, E.; et al. Massive Transient Damage of the Olfactory Epithelium Associated with Infection of Sustentacular Cells by SARS-CoV-2 in Golden Syrian Hamsters. Brain Behav. Immun. 2020, 89, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Shen, W.; Rowan, N.R.; Kulaga, H.; Hillel, A.; Ramanathan, M.; Lane, A.P. Elevated ACE-2 Expression in the Olfactory Neuroepithelium: Implications for Anosmia and Upper Respiratory SARS-CoV-2 Entry and Replication. Eur. Respir. J. 2020, 56, 2001948. [Google Scholar] [CrossRef]
- Fodoulian, L.; Tuberosa, J.; Rossier, D.; Boillat, M.; Kan, C.; Pauli, V.; Egervari, K.; Lobrinus, J.A.; Landis, B.N.; Carleton, A.; et al. SARS-CoV-2 Receptors and Entry Genes Are Expressed in the Human Olfactory Neuroepithelium and Brain. iScience 2020, 23, 101839. [Google Scholar] [CrossRef]
- Brann, D.H.; Tsukahara, T.; Weinreb, C.; Lipovsek, M.; van den Berge, K.; Gong, B.; Chance, R.; Macaulay, I.C.; Chou, H.J.; Fletcher, R.B.; et al. Non-Neuronal Expression of SARS-CoV-2 Entry Genes in the Olfactory System Suggests Mechanisms Underlying COVID-19-Associated Anosmia. Sci. Adv. 2020, 6, eabc5801. [Google Scholar] [CrossRef]
- Bilinska, K.; Jakubowska, P.; von Bartheld, C.S.; Butowt, R. Expression of the SARS-CoV-2 Entry Proteins, ACE2 and TMPRSS2, in Cells of the Olfactory Epithelium: Identification of Cell Types and Trends with Age. ACS Chem. Neurosci. 2020, 11, 1555–1562. [Google Scholar] [CrossRef]
- Divani, A.A.; Andalib, S.; Biller, J.; Napoli, D.M.; Moghimi, N.; Rubinos, C.A.; Nobleza, C.O.; Sylaja, P.N.; Toledano, M.; Lattanzi, S.; et al. Central Nervous System Manifestations Associated with COVID-19. Curr. Neurol. Neurosci. Rep. 2020, 20, 1–20. [Google Scholar] [CrossRef]
- Alipoor, S.D.; Mirsaeidi, M. SARS-CoV-2 Cell Entry beyond the ACE2 Receptor. Mol. Biol. Rep. 2022, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 Is a Host Factor for SARS-CoV-2 Infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 Facilitates SARS-CoV-2 Cell Entry and Infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Mayi, B.S.; Leibowitz, J.A.; Woods, A.T.; Ammon, K.A.; Liu, A.E.; Raja, A. The Role of Neuropilin-1 in COVID-19. PLoS Pathog. 2021, 17, e1009153. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Baker, S.A.; Kwok, S.; Berry, G.J.; Montine, T.J. Angiotensin-Converting Enzyme 2 (ACE2) Expression Increases with Age in Patients Requiring Mechanical Ventilation. PLoS ONE 2021, 16, e0247060. [Google Scholar] [CrossRef] [PubMed]
- Muus, C.; Luecken, M.D.; Eraslan, G.; Sikkema, L.; Waghray, A.; Heimberg, G.; Kobayashi, Y.; Vaishnav, E.D.; Subramanian, A.; Smillie, C.; et al. Single-Cell Meta-Analysis of SARS-CoV-2 Entry Genes across Tissues and Demographics. Nat. Med. 2021, 27, 546–559. [Google Scholar] [CrossRef]
- Ueha, R.; Kondo, K.; Kagoya, R.; Shichino, S.; Ueha, S.; Yamasoba, T. Ace2, Tmprss2, and Furin Expression in the Nose and Olfactory Bulb in Mice and Humans. Rhinology 2021, 59, 1–7. [Google Scholar] [CrossRef]
- Ziegler, C.G.K.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035.e19. [Google Scholar] [CrossRef]
- de Melo, G.D.; Lazarini, F.; Levallois, S.; Hautefort, C.; Michel, V.; Larrous, F.; Verillaud, B.; Aparicio, C.; Wagner, S.; Gheusi, G.; et al. COVID-19-Related Anosmia Is Associated with Viral Persistence and Inflammation in Human Olfactory Epithelium and Brain Infection in Hamsters. Sci. Transl. Med. 2021, 13, eabf8396. [Google Scholar] [CrossRef]
- Ye, Q.; Zhou, J.; He, Q.; Li, R.T.; Yang, G.; Zhang, Y.; Wu, S.J.; Chen, Q.; Shi, J.H.; Zhang, R.R.; et al. SARS-CoV-2 Infection in the Mouse Olfactory System. Cell Discov 2021, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Pekosz, A.; Villano, J.S.; Shen, W.; Zhou, R.; Kulaga, H.; Li, Z.; Beck, S.E.; Witwer, K.W.; Mankowski, J.L.; et al. Evolution of Nasal and Olfactory Infection Characteristics of SARS-CoV-2 Variants. bioRxiv 2022. [Google Scholar] [CrossRef]
- Armando, F.; Beythien, G.; Kaiser, F.K.; Allnoch, L.; Heydemann, L.; Rosiak, M.; Becker, S.; Gonzalez-Hernandez, M.; Lamers, M.M.; Haagmans, B.L.; et al. SARS-CoV-2 Omicron Variant Causes Mild Pathology in the Upper and Lower Respiratory Tract of Hamsters. Nat. Commun. 2022, 13, 3519. [Google Scholar] [CrossRef]
- Jia, C.; Roman, C.; Hegg, C.C. Nickel Sulfate Induces Location-Dependent Atrophy of Mouse Olfactory Epithelium: Protective and Proliferative Role of Purinergic Receptor Activation. Toxicol. Sci. 2010, 115, 547–556. [Google Scholar] [CrossRef]
- Heydel, J.M.; Coelho, A.; Thiebaud, N.; Legendre, A.; le Bon, A.M.; Faure, P.; Neiers, F.; Artur, Y.; Golebiowski, J.; Briand, L. Odorant-Binding Proteins and Xenobiotic Metabolizing Enzymes: Implications in Olfactory Perireceptor Events. ANat. Rec. 2013, 296, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhuang, Z.; Zheng, J.; Li, K.; Wong, R.L.Y.; Liu, D.; Huang, J.; He, J.; Zhu, A.; Zhao, J.; et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell 2020, 182, 734–743.e5. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T. Neurological Infection with SARS-CoV-2—The Story so Far. Nat. Rev. Neurol. 2021, 17, 65–66. [Google Scholar] [CrossRef]
- Vogalis, F.; Hegg, C.C.; Lucero, M.T. Ionic Conductances in Sustentacular Cells of the Mouse Olfactory Epithelium. J. Physiol. 2005, 562, 785–799. [Google Scholar] [CrossRef]
- Zazhytska, M.; Kodra, A.; Hoagland, D.A.; Frere, J.; Fullard, J.F.; Shayya, H.; McArthur, N.G.; Moeller, R.; Uhl, S.; Omer, A.D.; et al. Non-Cell-Autonomous Disruption of Nuclear Architecture as a Potential Cause of COVID-19-Induced Anosmia. Cell 2022, 185, 1052–1064.e12. [Google Scholar] [CrossRef]
- Shelton, J.F.; Shastri, A.J.; Fletez-Brant, K.; Auton, A.; Chubb, A.; Fitch, A.; Kung, A.; Altman, A.; Kill, A.; Shastri, A.J.; et al. The UGT2A1/UGT2A2 Locus Is Associated with COVID-19-Related Loss of Smell or Taste. Nat. Genet. 2022, 54, 121–124. [Google Scholar] [CrossRef]
- Finlay, J.B.; Brann, D.H.; Abi-Hachem, R.; Jang, D.W.; Oliva, A.D.; Ko, T.; Gupta, R.; Wellford, S.A.; Moseman, E.A.; Jang, S.S.; et al. Persistent Post-COVID-19 Smell Loss Is Associated with Inflammatory Infiltration and Altered Olfactory Epithelial Gene Expression. bioRxiv 2022. [Google Scholar] [CrossRef]
- Herrick, D.B.; Lin, B.; Peterson, J.; Schnittke, N.; Schwob, J.E. Notch1 Maintains Dormancy of Olfactory Horizontal Basal Cells, a Reserve Neural Stem Cell. Proc Natl. Acad Sci. USA 2017, 114, E5589–E5598. [Google Scholar] [CrossRef] [PubMed]
- Bergström, U.; Giovanetti, A.; Piras, E.; Brittebo, E.B. Methimazole-Induced Damage in the Olfactory Mucosa: Effects on Ultrastructure and Glutathione Levels. Toxicol. Pathol. 2003, 31, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Meinhardt, J.; Radke, J.; Dittmayer, C.; Franz, J.; Thomas, C.; Mothes, R.; Laue, M.; Schneider, J.; Brünink, S.; Greuel, S.; et al. Olfactory Transmucosal SARS-CoV-2 Invasion as a Port of Central Nervous System Entry in Individuals with COVID-19. Nat. Neurosci. 2021, 24, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.J.; Lee, A.C.Y.; Chu, H.; Chan, J.F.W.; Fan, Z.; Li, C.; Liu, F.; Chen, Y.; Yuan, S.; Poon, V.K.M.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 Infects and Damages the Mature and Immature Olfactory Sensory Neurons of Hamsters. Clin. Infect. Dis. 2021, 73, E503–E512. [Google Scholar] [CrossRef] [PubMed]
- Urata, S.; Maruyama, J.; Kishimoto-Urata, M.; Sattler, R.A.; Cook, R.; Lin, N.; Yamasoba, T.; Makishima, T.; Paessler, S. Regeneration Profiles of Olfactory Epithelium after SARS-CoV-2 Infection in Golden Syrian Hamsters. ACS Chem. Neurosci. 2021, 12, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Schwob, J.E.; Jang, W.; Holbrook, E.H.; Lin, B.; Herrick, D.B.; Peterson, J.N.; Hewitt Coleman, J. Stem and Progenitor Cells of the Mammalian Olfactory Epithelium: Taking Poietic License. J. Comp. Neurol. 2017, 525, 1034–1054. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, R.B.; Das, D.; Gadye, L.; Street, K.N.; Baudhuin, A.; Wagner, A.; Cole, M.B.; Flores, Q.; Choi, Y.G.; Yosef, N.; et al. Deconstructing Olfactory Stem Cell Trajectories at Single-Cell Resolution. Cell Stem. Cell 2017, 20, 817–830.e8. [Google Scholar] [CrossRef] [PubMed]
- Choi, R.; Goldstein, B.J. Olfactory Epithelium: Cells, Clinical Disorders, and Insights from an Adult Stem Cell Niche. Laryngoscope Investig. Otolaryngol. 2018, 3, 35–42. [Google Scholar] [CrossRef]
- Killingley, B.; Mann, A.J.; Kalinova, M.; Boyers, A.; Goonawardane, N.; Zhou, J.; Lindsell, K.; Hare, S.S.; Brown, J.; Frise, R.; et al. Safety, Tolerability and Viral Kinetics during SARS-CoV-2 Human Challenge in Young Adults. Nat. Med. 2022, 28, 1031–1041. [Google Scholar] [CrossRef]
- Sia, S.F.; Yan, L.M.; Chin, A.W.H.; Fung, K.; Choy, K.T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.A.P.M.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and Transmission of SARS-CoV-2 in Golden Hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Baig, A.M.; Khaleeq, A.; Ali, U.; Syeda, H. Evidence of the COVID-19 Virus Targeting the CNS: Tissue Distribution, Host-Virus Interaction, and Proposed Neurotropic Mechanisms. ACS Chem. Neurosci. 2020, 11, 995–998. [Google Scholar] [CrossRef] [PubMed]
- Schwob, J.E. Neural Regeneration and the Peripheral Olfactory System. Anat. Rec. 2002, 269, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Schwob, J.E.; Youngentob, S.L.; Mezza, R.C. Reconstitution of the Rat Olfactory Epithelium after Methyl Bromide-induced Lesion. J. Comp. Neurol. 1995, 359, 15–37. [Google Scholar] [CrossRef] [PubMed]
- Brann, J.H.; Firestein, S.J. A Lifetime of Neurogenesis in the Olfactory System. Front Neurosci 2014, 8, 182. [Google Scholar] [CrossRef] [PubMed]
- Liang, F. Sustentacular Cell Enwrapment of Olfactory Receptor Neuronal Dendrites: An Update. Genes 2020, 11, 493. [Google Scholar] [CrossRef] [PubMed]
- Chapurin, N.; Totten, D.J.; Chaballout, B.; Brennan, J.; Dennis, S.; Lubner, R.; Chowdhury, N.I.; Turner, J.H.; Trone, T.; Chandra, R.K. Differential Olfactory Outcomes in COVID-19: A Large Healthcare System Population Study. Int. Forum. Allergy Rhinol. 2022, 12, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, D.; Lahiri, B.; Ranjan, P.; Chatterjee, J.; Lahiri, P.; Sengupta, S. Road Map to Understanding Sars-Cov-2 Clinico-Immunopathology and Covid-19 Disease Severity. Pathogens 2021, 10, 5. [Google Scholar] [CrossRef]
- Saniasiaya, J.; Islam, M.A.; Abdullah, B. Prevalence of Olfactory Dysfunction in Coronavirus Disease 2019 (COVID-19): A Meta-Analysis of 27,492 Patients. Laryngoscope 2021, 131, 865–878. [Google Scholar] [CrossRef] [PubMed]
- Ziuzia-Januszewska, L.; Januszewski, M. Pathogenesis of Olfactory Disorders in COVID-19. Brain Sci. 2022, 12, 449. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahbaz, M.A.; De Bernardi, F.; Alatalo, A.; Sachana, M.; Clerbaux, L.-A.; Muñoz, A.; Parvatam, S.; Landesmann, B.; Kanninen, K.M.; Coecke, S. Mechanistic Understanding of the Olfactory Neuroepithelium Involvement Leading to Short-Term Anosmia in COVID-19 Using the Adverse Outcome Pathway Framework. Cells 2022, 11, 3027. https://doi.org/10.3390/cells11193027
Shahbaz MA, De Bernardi F, Alatalo A, Sachana M, Clerbaux L-A, Muñoz A, Parvatam S, Landesmann B, Kanninen KM, Coecke S. Mechanistic Understanding of the Olfactory Neuroepithelium Involvement Leading to Short-Term Anosmia in COVID-19 Using the Adverse Outcome Pathway Framework. Cells. 2022; 11(19):3027. https://doi.org/10.3390/cells11193027
Chicago/Turabian StyleShahbaz, Muhammad Ali, Francesca De Bernardi, Arto Alatalo, Magdalini Sachana, Laure-Alix Clerbaux, Amalia Muñoz, Surat Parvatam, Brigitte Landesmann, Katja M. Kanninen, and Sandra Coecke. 2022. "Mechanistic Understanding of the Olfactory Neuroepithelium Involvement Leading to Short-Term Anosmia in COVID-19 Using the Adverse Outcome Pathway Framework" Cells 11, no. 19: 3027. https://doi.org/10.3390/cells11193027
APA StyleShahbaz, M. A., De Bernardi, F., Alatalo, A., Sachana, M., Clerbaux, L. -A., Muñoz, A., Parvatam, S., Landesmann, B., Kanninen, K. M., & Coecke, S. (2022). Mechanistic Understanding of the Olfactory Neuroepithelium Involvement Leading to Short-Term Anosmia in COVID-19 Using the Adverse Outcome Pathway Framework. Cells, 11(19), 3027. https://doi.org/10.3390/cells11193027