Hepatic Peroxisome Proliferator-Activated Receptor Alpha Dysfunction in Porcine Septic Shock
Abstract
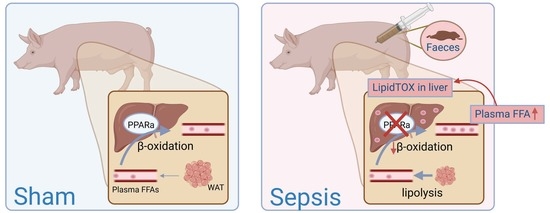
:1. Introduction
2. Materials and Methods
2.1. Experimental Procedure
2.2. Biological Samples
2.3. Liver Transcriptomic Analysis
2.4. LipidTOX Staining
2.5. Datasets and Databases
2.6. Statistics
3. Results
3.1. Hepatic PPARα Dysfunction in Porcine Septic Shock
3.1.1. Inflammation and Metabolic Dysregulation in Liver upon Septic Shock
3.1.2. PPARα Dysfunction
3.1.3. Increased FFA in Blood and Ectopic Lipid Accumulation in Liver
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Singer, M.; Deutschman, C.S.; Seymour, C.; Shankar-Har, M.; Annane, D.; Angus, D.C. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.v.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, Regional, and National Sepsis Incidence and Mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavaillon, J.; Singer, M.; Skirecki, T. Sepsis Therapies: Learning from 30 Years of Failure of Translational Research to Propose New Leads. EMBO Mol. Med. 2020, 12, e10128. [Google Scholar] [CrossRef] [PubMed]
- van Wyngene, L.; Vandewalle, J.; Libert, C. Reprogramming of Basic Metabolic Pathways in Microbial Sepsis: Therapeutic Targets at Last? EMBO Mol. Med. 2018, 10, e8712. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; Mcintyre, L.; Ostermann, M.; Prescott, H.C.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock 2021. Intensive Care Med. 2021, 47, 1181–1247. [Google Scholar] [CrossRef]
- Vandewalle, J.; Libert, C. Sepsis: A Failing Starvation Response. Trends Endocrinol. Metab. 2022, 33, 292–304. [Google Scholar] [CrossRef]
- Goldstein, I.; Baek, S.; Presman, D.M.; Paakinaho, V.; Swinstead, E.E.; Hager, G.L. Transcription Factor Assisted Loading and Enhancer Dynamics Dictate the Hepatic Fasting Response. Genome Res. 2017, 27, 427–439. [Google Scholar] [CrossRef] [Green Version]
- Kersten, S.; Stienstra, R. The Role and Regulation of the Peroxisome Proliferator Activated Receptor Alpha in Human Liver. Biochimie 2017, 136, 75–84. [Google Scholar] [CrossRef]
- Wang, A.; Huen, S.C.; Luan, H.H.; Yu, S.; Zhang, C.; Gallezot, J.-D.D.; Booth, C.J.; Medzhitov, R. Opposing Effects of Fasting Metabolism on Tissue Tolerance in Bacterial and Viral Inflammation. Cell 2016, 166, 1512–1525.e12. [Google Scholar] [CrossRef] [Green Version]
- Peterson, S.J.; Tsai, A.A.; Scala, C.M.; Sowa, D.C.; Sheean, P.M.; Braunschweig, C.L. Adequacy of Oral Intake in Critically Ill Patients 1 Week after Extubation. J. Am. Diet. Assoc. 2010, 110, 427–433. [Google Scholar] [CrossRef]
- Colaço, H.; Barros, A.; Neves-Costa, A.; Seixas, E.; Pedroso, D.; Velho, T.; Willmann, K.; Yi, H.-S.; Shong, M.; Benes, V.; et al. Host-Dependent Induction of Disease Tolerance to Infection by Tetracycline Antibiotics. Immunity 2020, 12, 53–67. [Google Scholar] [CrossRef]
- van Wyngene, L.; Vanderhaeghen, T.; Timmermans, S.; Vandewalle, J.; van Looveren, K.; Souffriau, J.; Wallaeys, C.; Eggermont, M.; Ernst, S.; van Hamme, E.; et al. Hepatic PPARα Function and Lipid Metabolic Pathways Are Dysregulated in Polymicrobial Sepsis. EMBO Mol. Med. 2020, 12, e11319. [Google Scholar] [CrossRef] [PubMed]
- Zingarelli, B.; Coopersmith, C.M.; Drechsler, S.; Efron, P.; Marshall, J.C.; Moldawer, L.; Wiersinga, W.J.; Xiao, X.; Osuchowski, M.F.; Thiemermann, C. Part I: Minimum Quality Threshold in Preclinical Sepsis Studies (MQTiPSS) for Study Design and Humane Modeling Endpoints. Shock 2019, 51, 10. [Google Scholar] [CrossRef] [PubMed]
- Guillon, A.; Preau, S.; Aboab, J.; Azabou, E.; Jung, B.; Silva, S.; Textoris, J.; Uhel, F.; Vodovar, D.; Zafrani, L.; et al. Preclinical Septic Shock Research: Why We Need an Animal ICU. Ann. Intensive Care 2019, 9, 66. [Google Scholar] [CrossRef] [Green Version]
- Bassols, A.; Costa, C.; Eckersall, P.D.; Osada, J.; Sabrià, J.; Tibau, J. The Pig as an Animal Model for Human Pathologies: A Proteomics Perspective. Proteom. Clin. Appl. 2014, 8, 715–731. [Google Scholar] [CrossRef]
- Meurens, F.; Summerfield, A.; Nauwynck, H.; Saif, L.; Gerdts, V. The Pig: A Model for Human Infectious Diseases. Trends Microbiol. 2012, 20, 50–57. [Google Scholar] [CrossRef]
- Garcia, B.; Su, F.; Dewachter, L.; Favory, R.; Khaldi, A.; Moiroux-Sahraoui, A.; Annoni, F.; Vasques-Nóvoa, F.; Rocha-Oliveira, E.; Roncon-Albuquerque, R.; et al. Myocardial Effects of Angiotensin II Compared to Norepinephrine in an Animal Model of Septic Shock. Crit. Care 2022, 26, 281. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime Cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.v.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, R.; Renqi, Y.; Lixue, W.; Xianzhong, X.; Yongming, Y. Minimum Quality Threshold in Pre-Clinical Sepsis Studies (MQTiPSS): Quality Thresholds for Study Design and Humane Modeling Endpoints. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 2019, 31, 1061–1071. [Google Scholar]
- Langley, R.J.; Tsalik, E.L.; Velkinburgh, J.C.V.; Glickman, S.W.; Rice, B.J.; Wang, C.; Chen, B.; Carin, L.; Suarez, A.; Mohney, R.P.; et al. An Integrated Clinico-Metabolomic Model Improves Prediction of Death in Sepsis. Sci. Transl. Med. 2013, 5, 195ra95. [Google Scholar] [CrossRef] [Green Version]
- Finn, P.F.; Dice, J.F. Proteolytic and Lipolytic Responses to Starvation. Nutrition 2006, 22, 830–844. [Google Scholar] [CrossRef] [PubMed]
- Vandewalle, J.; Timmermans, S.; Paakinaho, V.; Vancraeynest, L.; Dewyse, L.; Vanderhaeghen, T.; Wallaeys, C.; van Wyngene, L.; van Looveren, K.; Nuyttens, L.; et al. Combined Glucocorticoid Resistance and Hyperlactatemia Contributes to Lethal Shock in Sepsis. Cell Metab. 2021, 33, 1763–1776.e5. [Google Scholar] [CrossRef]
- Paumelle, R.; Haas, J.T.; Hennuyer, N.; Baugé, E.; Deleye, Y.; Mesotten, D.; Langouche, L.; Vanhoutte, J.; Cudejko, C.; Wouters, K.; et al. Hepatic PPARα Is Critical in the Metabolic Adaptation to Sepsis. J. Hepatol. 2019, 70, 963–973. [Google Scholar] [CrossRef]
- Wong, H.R.; Cvijanovich, N.; Allen, G.L.; Lin, R.; Anas, N.; Meyer, K.; Freishtat, R.J.; Monaco, M.; Odoms, K.; Sakthivel, B.; et al. Genomic Expression Profiling across the Pediatric Systemic Inflammatory Response Syndrome, Sepsis, and Septic Shock Spectrum. Crit. Care Med. 2009, 37, 1558. [Google Scholar] [CrossRef] [Green Version]
- van der Linden, A.; Blokker, B.M.; Kap, M.; Weustink, A.C.; Riegman, P.H.J.; Oosterhuis, J.W.; Cappello, F. Post-Mortem Tissue Biopsies Obtained at Minimally Invasive Autopsy: An RNA-Quality Analysis. PLoS ONE 2014, 9, e115675. [Google Scholar] [CrossRef] [Green Version]
- van Malenstein, H.; Wauters, J.; Mesotten, D.; Langouche, L.; de Vos, R.; Wilmer, A.; van Pelt, J. Molecular Analysis of Sepsis-Induced Changes in the Liver: Microarray Study in a Porcine Model of Acute Fecal Peritonitis with Fluid Resuscitation. Shock 2010, 34, 427–436. [Google Scholar] [CrossRef]




| Sham | Septic Shock | p-Value | ||
|---|---|---|---|---|
| Hemodynamic parameters | MAP (mmHg) | 68 ± 2 | 68 ± 3 | 0.85 |
| HR (/min) | 87 ± 3 | 155 ± 21 | 0.03 | |
| CO (L/min) | 6 ± 1 | 9 ± 2 | 0.03 | |
| NE (μg/kg/min) | 0 ± 0 | 1 ± 0.84 | 0.08 | |
| SvO2 (%) | 64 ± 3 | 73 ± 5 | 0.02 | |
| PCO2 gap (mmHg) | 5 ± 1 | 5 ± 5 | 0.98 | |
| Respiratory function | RR (/min) | 17 ± 3 | 20 ± 3 | 0.10 |
| Tidal Volume (mL) | 375 ± 35 | 385 ± 50 | 0.76 | |
| PaCO2 (mm Hg) | 52 ± 1 | 47 ± 5 | 0.15 | |
| PaO2/FiO2 ratio | 330 ± 58 | 253 ± 58 | 0.07 | |
| Metabolism | T° | 38.8 ± 0.8 | 38.8 ± 0.4 | 0.80 |
| Glucose (mg/dL) | 107 ± 10 | 115 ± 23 | 0.61 | |
| Lactate (mmol/L) | 0.9 ± 0.00 | 2 ± 1.1 | 0.12 | |
| pH | 7.46 ± 0.01 | 7.42 ± 0.06 | 0.26 | |
| Base excess (mmol/L) | 12 ± 1 | 5 ± 5 | 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vandewalle, J.; Garcia, B.; Timmermans, S.; Vanderhaeghen, T.; Van Wyngene, L.; Eggermont, M.; Dufoor, H.; Van Dender, C.; Halimi, F.; Croubels, S.; et al. Hepatic Peroxisome Proliferator-Activated Receptor Alpha Dysfunction in Porcine Septic Shock. Cells 2022, 11, 4080. https://doi.org/10.3390/cells11244080
Vandewalle J, Garcia B, Timmermans S, Vanderhaeghen T, Van Wyngene L, Eggermont M, Dufoor H, Van Dender C, Halimi F, Croubels S, et al. Hepatic Peroxisome Proliferator-Activated Receptor Alpha Dysfunction in Porcine Septic Shock. Cells. 2022; 11(24):4080. https://doi.org/10.3390/cells11244080
Chicago/Turabian StyleVandewalle, Jolien, Bruno Garcia, Steven Timmermans, Tineke Vanderhaeghen, Lise Van Wyngene, Melanie Eggermont, Hester Dufoor, Céline Van Dender, Fëllanza Halimi, Siska Croubels, and et al. 2022. "Hepatic Peroxisome Proliferator-Activated Receptor Alpha Dysfunction in Porcine Septic Shock" Cells 11, no. 24: 4080. https://doi.org/10.3390/cells11244080
APA StyleVandewalle, J., Garcia, B., Timmermans, S., Vanderhaeghen, T., Van Wyngene, L., Eggermont, M., Dufoor, H., Van Dender, C., Halimi, F., Croubels, S., Herpain, A., & Libert, C. (2022). Hepatic Peroxisome Proliferator-Activated Receptor Alpha Dysfunction in Porcine Septic Shock. Cells, 11(24), 4080. https://doi.org/10.3390/cells11244080