Histone Chaperone Nrp1 Mutation Affects the Acetylation of H3K56 in Tetrahymena thermophila
Abstract
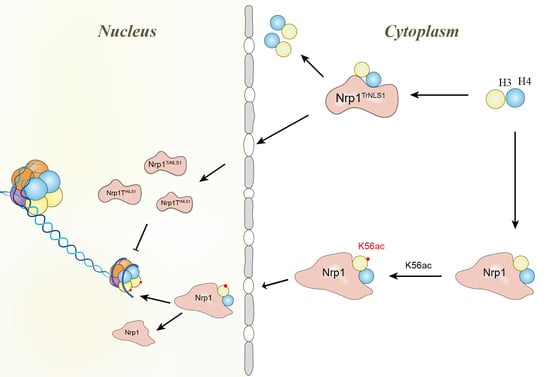
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture
2.2. Protein HA Tagging
2.3. RNA Extraction and qRT-PCR
2.4. Indirect Immunofluorescence
2.5. Histone Isolation
2.6. Western Blotting
2.7. Micronuclear Integrity Assay
2.8. Flow Cytometry
2.9. RNA Sequencing and Bioinformatic Analysis
3. Results
3.1. Characterization of Nrp1 Nuclear Localization Signals
3.2. Nrp1 Mutation Affects Chromatin Stability
3.3. Overexpression of Nrp1TrNLS1 Abolishes Histone H3 Nuclear Import
3.4. Modification of H3K56 Acetylation Decreases in Nrp1 Mutants
3.5. Nrp1TrNLS1 Mutants Affect the MIC Structure during Sexual Development
3.6. Nrp1TrNLS1 Mutant Affects Macronuclear Genome Transcription
4. Discussion
4.1. NLS1 or NLS2 Is Sufficient for Directing Nrp1 Nuclear Import
4.2. Nrp1 Mutation Leads to Nuclear Degradation and Cellular Apoptosis
4.3. Nrp1 Mutations Affect Genomic Transcription
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Akey, C.W.; Luger, K. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 2003, 13, 6–14. [Google Scholar] [CrossRef]
- Bernardes, N.E.; Chook, Y.M. Nuclear import of histones. Biochem. Soc. Trans. 2020, 48, 2753–2767. [Google Scholar] [CrossRef] [PubMed]
- De Koning, L.; Corpet, A.; Haber, J.E.; Almouzni, G. Histone chaperones: An escort network regulating histone traffic. Nat. Struct. Mol. Biol. 2007, 14, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Das, C.; Tyler, J.K.; Churchill, M.E. The histone shuffle: Histone chaperones in an energetic dance. Trends Biochem. Sci. 2010, 35, 476–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahlin, J.L.; Chen, X.; Walters, M.A.; Zhang, Z. Histone-modifying enzymes, histone modifications and histone chaperones in nucleosome assembly: Lessons learned from Rtt109 histone acetyltransferases. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 31–53. [Google Scholar] [CrossRef] [Green Version]
- Haushalter, K.A.; Kadonaga, J.T. Chromatin assembly by DNA-translocating motors. Nat. Rev. Mol. Cell Biol. 2003, 4, 613–620. [Google Scholar] [CrossRef]
- Kim, H.J.; Seol, J.H.; Han, J.W.; Youn, H.D.; Cho, E.J. Histone chaperones regulate histone exchange during transcription. EMBO J. 2007, 26, 4467–4474. [Google Scholar] [CrossRef] [Green Version]
- Tagami, H.; Ray-Gallet, D.; Almouzni, G.; Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 2004, 116, 51–61. [Google Scholar] [CrossRef] [Green Version]
- Drane, P.; Ouararhni, K.; Depaux, A.; Shuaib, M.; Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010, 24, 1253–1265. [Google Scholar] [CrossRef] [Green Version]
- Campos, E.I.; Fillingham, J.; Li, G.; Zheng, H.; Voigt, P.; Kuo, W.H.; Seepany, H.; Gao, Z.; Day, L.A.; Greenblatt, J.F.; et al. The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 2010, 17, 1343–1351. [Google Scholar] [CrossRef] [Green Version]
- Parthun, M.R.; Widom, J.; Gottschling, D.E. The major cytoplasmic histone acetyltransferase in yeast: Links to chromatin replication and histone metabolism. Cell 1996, 87, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Recht, J.; Tsubota, T.; Tansy, J.C.; Diaz, R.L.; Berger, J.M.; Zhang, X.; Garcia, B.A.; Shabanowitzn, J.; Burlingame, A.L.; Hunt, D.F.; et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 2006, 103, 6988–6993. [Google Scholar] [CrossRef] [Green Version]
- Xhemalce, B.; Miller, K.M.; Driscoll, R.; Masumoto, H.; Jackson, S.P.; Kouzarides, T.; Verreault, A.; Arcangioli, B. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J. Biol. Chem. 2007, 282, 15040–15047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topal, S.; Vasseur, P.; Radman-Livaja, M.; Peterson, C.L. Distinct transcriptional roles for Histone H3-K56 acetylation during the cell cycle in Yeast. Nat. Commun. 2019, 10, 4372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, C.; Lucia, M.S.; Hansen, K.C.; Tyler, J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 2009, 459, 113–117. [Google Scholar] [CrossRef]
- Li, Y.; Jaramillo-Lambert, A.N.; Yang, Y.; Williams, R.; Lee, N.H.; Zhu, W. And-1 is required for the stability of histone acetyltransferase Gcn5. Oncogene 2012, 31, 643–652. [Google Scholar] [CrossRef] [Green Version]
- Filippakopoulos, P.; Picaud, S.; Mangos, M.; Keates, T.; Lambert, J.P.; Barsyte-Lovejoy, D.; Felletar, I.; Volkmer, R.; Muller, S.; Pawson, T.; et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149, 214–231. [Google Scholar] [CrossRef] [Green Version]
- Kuo, M.-H.; Brownell, J.E.; Sobel, R.E.; Ranalli, T.A.; Cook, R.G.; Edmondson, D.G.; Roth, S.Y.; Allis, C.D. Transcription-linked acetylation by GcnSp of histones H3 and H4 at specific lysines. Nature 1996, 383, 269–272. [Google Scholar] [CrossRef]
- Burgess, R.J.; Zhou, H.; Han, J.; Zhang, Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol. Cell. 2010, 37, 469–480. [Google Scholar] [CrossRef] [Green Version]
- Stejskal, S.; Stepka, K.; Tesarova, L.; Stejskal, K.; Matejkova, M.; Simara, P.; Zdrahal, Z.; Koutna, I. Cell cycle-dependent changes in H3K56ac in human cells. Cell Cycle 2015, 14, 3851–3863. [Google Scholar] [CrossRef]
- Orias, E.; Cervantes, M.D.; Hamilton, E.P. Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res. Microbiol. 2011, 162, 578–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, E.; Sugai, T. Developmental progression of Tetrahymena through the cell cycle and conjugation. Methods Cell Biol. 2012, 109, 177–236. [Google Scholar] [PubMed]
- Karrer, K.K. Tetrahymena genetics: Two nuclei are better than one. J. Methods Cell Biol. 1999, 62, 127–186. [Google Scholar]
- Bednenko, J.; Noto, T.; DeSouza, L.V.; Siu, K.W.; Pearlman, R.E.; Mochizuki, K.; Gorovsky, M.A. Two GW repeat proteins interact with Tetrahymena thermophila argonaute and promote genome rearrangement. Mol. Cell Biol. 2009, 29, 5020–5030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, H.; Xu, J.; Wang, W. Ran1 is essential for parental macronuclear import of apoptosis-inducing factor and programmed nuclear death in Tetrahymena thermophila. FEBS J. 2019, 286, 913–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garg, J.; Lambert, J.P.; Karsou, A.; Marquez, S.; Nabeel-Shah, S.; Bertucci, V.; Retnasothie, D.V.; Radovani, E.; Pawson, T.; Gingras, A.C.; et al. Conserved Asf1-importin beta physical interaction in growth and sexual development in the ciliate Tetrahymena thermophila. J. Proteom. 2013, 94, 311–326. [Google Scholar] [CrossRef] [Green Version]
- Lian, Y.; Hao, H.; Xu, J.; Bo, T.; Liang, A.; Wang, W. The histone chaperone Nrp1 is required for chromatin stability and nuclear division in Tetrahymena thermophila. Epigenetics Chromatin 2021, 14, 34. [Google Scholar] [CrossRef]
- Gorovsky, M.A.; Yao, M.-C.; Keevert, J.B.; Pleger, G.L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975, 9, 311–327. [Google Scholar]
- Bruns, P.J.; Brussard, T.B. Pair formation in Tetrahymena pyriformis, an inducible developmental system. J. Exp. Zool. 1974, 188, 337–344. [Google Scholar] [CrossRef]
- Hamilton, E.; Bruns, P.; Lin, C.; Merriam, V.; Orias, E.; Vong, L.; Cassidy-Hanley, D. Genome-wide characterization of tetrahymena thermophila chromosome breakage sites. I. Cloning and identification of functional sites. Genetics 2005, 170, 1611–1621. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Li, H.; Zhang, J.; Tian, X. Effect of melamine toxicity on Tetrahymena thermophila proliferation and metallothionein expression. Food Chem. Toxicol. 2015, 80, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Dunleavy, E.M.; Pidoux, A.L.; Monet, M.; Bonilla, C.; Richardson, W.; Hamilton, G.L.; Ekwall, K.; McLaughlin, P.J.; Allshire, R.C. A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres. Mol. Cell. 2007, 28, 1029–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, J.; Xu, J.; Bo, T.; Wang, W. Micronucleus-specific histone H1 is required for micronuclear chromosome integrity in Tetrahymena thermophila. PLoS ONE 2017, 12, e0187475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nabeel-Shah, S.; Ashraf, K.; Pearlman, R.E.; Fillingham, J. Molecular evolution of NASP and conserved histone H3/H4 transport pathway. BMC Evol. Biol. 2014, 14, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skalska, L.; Stojnic, R.; Li, J.; Fischer, B.; Cerda-Moya, G.; Sakai, H.; Tajbakhsh, S.; Russell, S.; Adryan, B.; Bray, S.J. Chromatin signatures at Notch-regulated enhancers reveal large-scale changes in H3K56ac upon activation. EMBO J. 2015, 34, 1889–1904. [Google Scholar] [CrossRef]
- Wahab, S.; Saettone, A.; Nabeel-Shah, S.; Dannah, N.; Fillingham, J. Exploring the Histone Acetylation Cycle in the Protozoan Model Tetrahymena thermophila. Front Cell Dev Biol. 2020, 8, 509. [Google Scholar] [CrossRef]
- Han, J.; Zhou, H.; Li, Z.; Xu, R.M.; Zhang, Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J. Biol. Chem. 2007, 282, 28587–28596. [Google Scholar] [CrossRef] [Green Version]
- Wolfe, J.; Hunter, B.; Adair, W.S. A cytological study of micronuclear elongation during conjugation in Tetrahymena. Chromosoma 1976, 55, 289–308. [Google Scholar] [CrossRef]
- Sugai, T.; Hiwatashi, K. Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in Tetrahymena pyriformis. J. Eukaryot. Microbiol. 1974, 21, 542–548. [Google Scholar]
- Bowman, A.; Lercher, L.; Singh, H.R.; Zinne, D.; Timinszky, G.; Carlomagno, T.; Ladurner, A.G. The histone chaperone sNASP binds a conserved peptide motif within the globular core of histone H3 through its TPR repeats. Nucleic Acids Res. 2016, 44, 3105–3117. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Walsh, S.T.; Parthun, M.R. Expanded binding specificity of the human histone chaperone NASP. Nucleic Acids Res. 2008, 36, 5763–5772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osakabe, A.; Tachiwana, H.; Matsunaga, T.; Shiga, T.; Nozawa, R.S.; Obuse, C.; Kurumizaka, H. Nucleosome formation activity of human somatic nuclear autoantigenic sperm protein (sNASP). J. Biol. Chem. 2010, 285, 11913–11921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Ge, Z.; Walsh, S.T.; Parthun, M.R. The human histone chaperone sNASP interacts with linker and core histones through distinct mechanisms. Nucleic Acids Res. 2012, 40, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Alekseev, O.M.; Bencic, D.C.; Richardson, R.T.; Widgren, E.E.; O’Rand, M.G. Overexpression of the Linker histone-binding protein tNASP affects progression through the cell cycle. J. Biol. Chem. 2003, 278, 8846–8852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dannah, N.S.; Nabeel-Shah, S.; Kurat, C.F.; Sabatinos, S.A.; Fillingham, J. Functional Analysis of Hif1 Histone Chaperone in Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2018, 8, 1993–2006. [Google Scholar] [CrossRef] [Green Version]
- Groth, A.; Corpet, A.; Cook, A.J.L.; Roche, D.; Bartek, J.; Lukas, J.; Almouzni, G. Regulation of replication fork progression through histone supply and demand. Science 2007, 318, 1928–1931. [Google Scholar] [CrossRef]
- Jasencakova, Z.; Scharf, A.N.; Ask, K.; Corpet, A.; Imhof, A.; Almouzni, G.; Groth, A. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell. 2010, 37, 736–743. [Google Scholar] [CrossRef]
- Alekseev, O.M.; Richardson, R.T.; Alekseev, O.; O’Rand, M.G. Analysis of gene expression profiles in HeLa cells in response to overexpression or siRNA-mediated depletion of NASP. Reprod Biol Endocrinol. 2009, 7, 45. [Google Scholar] [CrossRef] [Green Version]
- Alekseev, O.M.; Richardson, R.T.; Tsuruta, J.K.; O’Rand, M.G. Depletion of the histone chaperone tNASP inhibits proliferation and induces apoptosis in prostate cancer PC-3 cells. Reprod. Biol. Endocrinol. 2011, 9, 50. [Google Scholar] [CrossRef] [Green Version]
- Kang, X.; Feng, Y.; Gan, Z.; Zeng, S.; Guo, X.; Chen, X.; Zhang, Y.; Wang, C.; Liu, K.; Chen, X.; et al. NASP antagonize chromatin accessibility through maintaining histone H3K9me1 in hepatocellular carcinoma. Biochim Biophys Acta Mol Basis Dis. 2018, 1864, 3438–3448. [Google Scholar] [CrossRef]
- Cook, A.J.L.; Gurard-Levin, Z.A.; Vassias, I.; Almouzni, G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol Cell. 2011, 44, 918–927. [Google Scholar] [CrossRef] [Green Version]
- Daganzo, S.M.; Erzberger, J.P.; Lam, W.M.; Skordalakes, E.; Zhang, R.; Franco, A.A.; Brill, S.J.; Adams, P.D.; Berger, J.M.; Kaufman, P.D. Structure and function of the conserved core of histone deposition protein Asf1. Curr Biol. 2003, 13, 2148–2158. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, A.J.; Dacks, J.B.; Field, M.C. Evolution of the karyopherin-beta family of nucleocytoplasmic transport factors; ancient origins and continued specialization. PLoS ONE 2011, 6, e19308. [Google Scholar]
- Nabeel-Shah, S.; Garg, J.; Saettone, A.; Ashraf, K.; Lee, H.; Wahab, S.; Ahmed, N.; Fine, J.; Derynck, J.; Pu, S.; et al. Functional characterization of RebL1 highlights the evolutionary conservation of oncogenic activities of the RBBP4/7 orthologue in Tetrahymena thermophila. Nucleic Acids Res. 2021, 49, 6196–6212. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, A.T.; Seale, R.L. Histone deacetylation is required for the maturation of newly replicated chromatin. J. Biol. Chem. 1983, 258, 12675–12684. [Google Scholar] [CrossRef]
- Shimko, J.C.; North, J.A.; Bruns, A.N.; Poirier, M.G.; Ottesen, J.J. Preparation of fully synthetic histone H3 reveals that acetyl-lysine 56 facilitates protein binding within nucleosomes. J. Mol. Biol. 2011, 408, 187–204. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Pu, M.T.; Zhang, Z.G.; Lou, Z.K. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle 2009, 8, 1747–1753. [Google Scholar] [CrossRef] [Green Version]
- Celic, I.; Masumoto, H.; Griffith, W.P.; Meluh, P.; Cotter, R.J.; Boeke, J.D.; Verreault, A. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr. Biol. 2006, 16, 1280–1289. [Google Scholar] [CrossRef] [Green Version]
- Garcia, B.A.; Hake, S.B.; Diaz, R.L.; Kauer, M.; Morris, S.A.; Recht, J.; Shabanowitz, J.; Mishra, N.; Strahl, B.D.; Allis, C.D.; et al. Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 2007, 282, 7641–7655. [Google Scholar] [CrossRef] [Green Version]
- Park, K.; Kim, J.A.; Kim, J. Transcriptional regulation by the KMT2 histone H3K4 methyltransferases. Biochim. Biophys. Acta Gene Regul Mech. 2020, 1863, 194545. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinheiro, I.; Margueron, R.; Shukeir, N.; Eisold, M.; Fritzsch, C.; Richter, F.M.; Mittler, G.; Genoud, C.; Goyama, S.; Kurokawa, M.; et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 2012, 150, 948–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Zhang, K.; Grunstein, M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 2005, 121, 375–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernier, M.; Luo, Y.; Nwokelo, K.C.; Goodwin, M.; Dreher, S.J.; Zhang, P.; Parthun, M.R.; Fondufe-Mittendorf, Y.; Ottesen, J.J.; Poirier, M.G. Linker histone H1 and H3K56 acetylation are antagonistic regulators of nucleosome dynamics. Nat. Commun. 2015, 6, 10152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; Xue, Y.; Song, C.; Grunstein, M. Acetylated histone H3K56 interacts with Oct4 to promote mouse embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. USA 2013, 110, 11493–11498. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Cao, J.; Wang, Y.; Xu, J.; Zhou, Z.; Gu, X.; Liu, X.; Wen, H.; Wu, H.; Cheng, C. Transcription initiation factor IIB involves in Schwann cell differentiation after rat sciatic nerve crush. J. Mol. Neurosci. 2013, 49, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Fant, C.B.; Levandowski, C.B.; Gupta, K.; Maas, Z.L.; Moir, J.; Rubin, J.D.; Sawyer, A.; Esbin, M.N.; Rimel, J.K.; Luyties, O.; et al. TFIID Enables RNA Polymerase II Promoter-Proximal Pausing. Mol. Cell 2020, 78, 785–793.e8. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lian, Y.; Hao, H.; Xu, J.; Bo, T.; Wang, W. Histone Chaperone Nrp1 Mutation Affects the Acetylation of H3K56 in Tetrahymena thermophila. Cells 2022, 11, 408. https://doi.org/10.3390/cells11030408
Lian Y, Hao H, Xu J, Bo T, Wang W. Histone Chaperone Nrp1 Mutation Affects the Acetylation of H3K56 in Tetrahymena thermophila. Cells. 2022; 11(3):408. https://doi.org/10.3390/cells11030408
Chicago/Turabian StyleLian, Yinjie, Huijuan Hao, Jing Xu, Tao Bo, and Wei Wang. 2022. "Histone Chaperone Nrp1 Mutation Affects the Acetylation of H3K56 in Tetrahymena thermophila" Cells 11, no. 3: 408. https://doi.org/10.3390/cells11030408
APA StyleLian, Y., Hao, H., Xu, J., Bo, T., & Wang, W. (2022). Histone Chaperone Nrp1 Mutation Affects the Acetylation of H3K56 in Tetrahymena thermophila. Cells, 11(3), 408. https://doi.org/10.3390/cells11030408