Functional and Genetic Characterization of Porcine Beige Adipocytes
Abstract
:1. Introduction
2. Materials and Methods
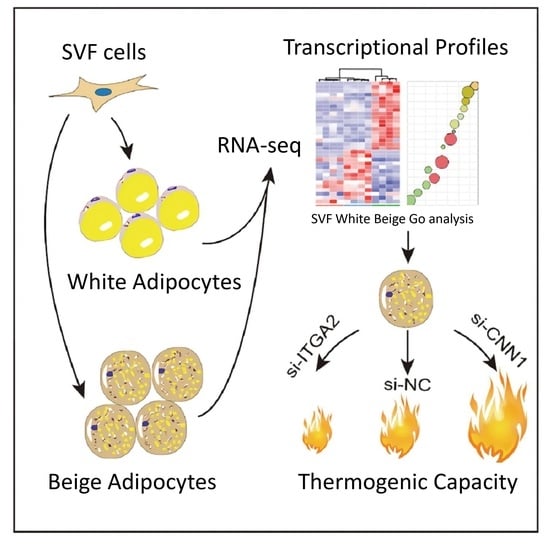
2.1. Isolation, Culture, and Differentiation of Porcine SVF Cells
2.2. Western Blot
2.3. RNA Isolation, RT–PCR, and QPCR Analysis
2.4. mRNA Sequencing, RNA-seq Data Analysis and Functional Analysis
2.5. Oil Red-O Staining
2.6. Fluorescence Microscopy
2.7. Image Acquisition and Processing
2.8. Seahorse Metabolic Assays
2.9. RNA Interference
2.10. Statistical Analysis
3. Results
3.1. The Establishment of an In Vitro Model of Porcine Beige Adipogenesis
3.2. Distinct Transcriptional Profiles of Porcine White and Beige Adipocytes
3.3. Identification of Beige Adipogenesis-Related Candidate Genes
3.4. Comparative Analysis of DEGs in Beige Adipogenesis
3.5. ITAG2 and CNN1 Are Involved in Porcine Beige Adipocyte Differentiation and Thermogenesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shao, M.; Wang, Q.A.; Song, A.; Vishvanath, L.; Busbuso, N.C.; Scherer, P.E.; Gupta, R.K. Cellular Origins of Beige Fat Cells Revisited. Diabetes 2019, 68, 1874–1885. [Google Scholar] [CrossRef]
- Ikeda, K.; Maretich, P.; Kajimura, S. The Common and Distinct Features of Brown and Beige Adipocytes. Trends Endocrinol. Metab. 2018, 29, 191–200. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.D.; Spiegelman, B.M. What we talk about when we talk about fat. Cell 2014, 156, 20–44. [Google Scholar] [CrossRef] [Green Version]
- Giralt, M.; Villarroya, F. White, brown, beige/brite: Different adipose cells for different functions? Endocrinology 2013, 154, 2992–3000. [Google Scholar] [CrossRef] [Green Version]
- Ahfeldt, T.; Schinzel, R.T.; Lee, Y.K.; Hendrickson, D.; Kaplan, A.; Lum, D.H.; Camahort, R.; Xia, F.; Shay, J.; Rhee, E.P.; et al. Programming human pluripotent stem cells into white and brown adipocytes. Nat. Cell. Biol. 2012, 14, 209–219. [Google Scholar] [CrossRef] [Green Version]
- Su, S.; Guntur, A.R.; Nguyen, D.C.; Fakory, S.S.; Doucette, C.C.; Leech, C.; Lotana, H.; Kelley, M.; Kohli, J.; Martino, J.; et al. A Renewable Source of Human Beige Adipocytes for Development of Therapies to Treat Metabolic Syndrome. Cell Rep. 2018, 25, 3215–3228 e3219. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.M.; Zhang, L.; Avery, J.; Yin, A.; Du, Y.; Wang, H.; Li, Z.; Fu, H.; Yin, H.; Dalton, S. Human beige adipocytes for drug discovery and cell therapy in metabolic diseases. Nat. Commun. 2020, 11, 2758. [Google Scholar] [CrossRef]
- Wu, J.; Bostrom, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef] [Green Version]
- Xue, R.; Lynes, M.D.; Dreyfuss, J.M.; Shamsi, F.; Schulz, T.J.; Zhang, H.; Huang, T.L.; Townsend, K.L.; Li, Y.; Takahashi, H.; et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat. Med. 2015, 21, 760–768. [Google Scholar] [CrossRef]
- Shinoda, K.; Luijten, I.H.; Hasegawa, Y.; Hong, H.; Sonne, S.B.; Kim, M.; Xue, R.; Chondronikola, M.; Cypess, A.M.; Tseng, Y.H.; et al. Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat. Med. 2015, 21, 389–394. [Google Scholar] [CrossRef] [Green Version]
- Ussar, S.; Lee, K.Y.; Dankel, S.N.; Boucher, J.; Haering, M.F.; Kleinridders, A.; Thomou, T.; Xue, R.; Macotela, Y.; Cypess, A.M.; et al. ASC-1, PAT2, and P2RX5 are cell surface markers for white, beige, and brown adipocytes. Sci. Transl. Med. 2014, 6, 247ra103. [Google Scholar] [CrossRef] [Green Version]
- Garcia, R.A.; Roemmich, J.N.; Claycombe, K.J. Evaluation of markers of beige adipocytes in white adipose tissue of the mouse. Nutr. Metab. 2016, 13, 24. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, T.; Sakai, J.; Kajimura, S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat. Rev. Mol. Cell. Biol. 2016, 17, 480–495. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Seale, P. Control of brown and beige fat development. Nat. Rev. Mol. Cell. Biol. 2016, 17, 691–702. [Google Scholar] [CrossRef]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell. Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef]
- Li, K.; Liu, C.; Wahlqvist, M.L.; Li, D. Econutrition, brown and beige fat tissue and obesity. Asia Pac. J. Clin. Nutr. 2020, 29, 668–680. [Google Scholar] [CrossRef]
- Nicholls, D.G. The hunt for the molecular mechanism of brown fat thermogenesis. Biochimie 2017, 134, 9–18. [Google Scholar] [CrossRef]
- Berg, F.; Gustafson, U.; Andersson, L. The uncoupling protein 1 gene (UCP1) is disrupted in the pig lineage: A genetic explanation for poor thermoregulation in piglets. PLoS Genet. 2006, 2, e129. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.; Lin, J.; Huang, J.; Zhang, H.; Zhang, R.; Zhang, X.; Cao, C.; Hambly, C.; Qin, G.; Yao, J.; et al. Reconstitution of UCP1 using CRISPR/Cas9 in the white adipose tissue of pigs decreases fat deposition and improves thermogenic capacity. Proc. Natl. Acad. Sci. USA 2017, 114, E9474–E9482. [Google Scholar] [CrossRef] [Green Version]
- Louveau, I.; Perruchot, M.H.; Bonnet, M.; Gondret, F. Invited review: Pre- and postnatal adipose tissue development in farm animals: From stem cells to adipocyte physiology. Animal 2016, 10, 1839–1847. [Google Scholar] [CrossRef]
- Lin, J.; Cao, C.; Tao, C.; Ye, R.; Dong, M.; Zheng, Q.; Wang, C.; Jiang, X.; Qin, G.; Yan, C.; et al. Cold adaptation in pigs depends on UCP3 in beige adipocytes. J. Mol. Cell. Biol. 2017, 9, 364.4–375. [Google Scholar] [CrossRef]
- Gao, Y.; Qimuge, N.R.; Qin, J.; Cai, R.; Li, X.; Chu, G.Y.; Pang, W.J.; Yang, G.S. Acute and chronic cold exposure differentially affects the browning of porcine white adipose tissue. Animal 2018, 12, 1435–1441. [Google Scholar] [CrossRef] [Green Version]
- Hou, L.; Xie, M.; Cao, L.; Shi, J.; Xu, G.; Hu, C.; Wang, C. Browning of Pig White Preadipocytes by Co-Overexpressing Pig PGC-1alpha and Mice UCP1. Cell Physiol. Biochem. 2018, 48, 556–568. [Google Scholar] [CrossRef]
- Benador, I.Y.; Veliova, M.; Mahdaviani, K.; Petcherski, A.; Wikstrom, J.D.; Assali, E.A.; Acin-Perez, R.; Shum, M.; Oliveira, M.F.; Cinti, S.; et al. Mitochondria Bound to Lipid Droplets Have Unique Bioenergetics, Composition, and Dynamics that Support Lipid Droplet Expansion. Cell Metab. 2018, 27, 869–885 e866. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Miao, Y.R.; Jia, L.H.; Yu, Q.Y.; Zhang, Q.; Guo, A.Y. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019, 47, D33–D38. [Google Scholar] [CrossRef]
- Wang, W.; Kissig, M.; Rajakumari, S.; Huang, L.; Lim, H.W.; Won, K.J.; Seale, P. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc. Natl. Acad. Sci. USA 2014, 111, 14466–14471. [Google Scholar] [CrossRef] [Green Version]
- Stine, R.R.; Shapira, S.N.; Lim, H.W.; Ishibashi, J.; Harms, M.; Won, K.J.; Seale, P. EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol. Metab. 2016, 5, 57–65. [Google Scholar] [CrossRef]
- Chen, Y.C.; Yu, Y.H. The potential of brown adipogenesis and browning in porcine bone marrow-derived mesenchymal stem cells1. J. Anim. Sci. 2018, 96, 3635–3644. [Google Scholar] [CrossRef]
- Castillo-Duran, C.; Vial, P.; Uauy, R. Oral copper supplementation: Effect on copper and zinc balance during acute gastroenteritis in infants. Am. J. Clin. Nutr. 1990, 51, 1088–1092. [Google Scholar] [CrossRef]
- Kim, K.; Nam, K.H.; Yi, S.A.; Park, J.W.; Han, J.W.; Lee, J. Ginsenoside Rg3 Induces Browning of 3T3-L1 Adipocytes by Activating AMPK Signaling. Nutrients 2020, 12, 427. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Galilea, M.; Perez-Matute, P.; Prieto-Hontoria, P.L.; Houssier, M.; Burrell, M.A.; Langin, D.; Martinez, J.A.; Moreno-Aliaga, M.J. alpha-Lipoic acid treatment increases mitochondrial biogenesis and promotes beige adipose features in subcutaneous adipocytes from overweight/obese subjects. Biochim. Biophys. Acta 2015, 1851, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, T.; Villareal, M.O.; Motojima, H.; Isoda, H. Increasing cAMP levels of preadipocytes by cyanidin-3-glucoside treatment induces the formation of beige phenotypes in 3T3-L1 adipocytes. J. Nutr. Biochem. 2017, 40, 77–85. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wei, Y.; Ning, C.; Zhang, M.; Fan, P.; Lei, D.; Du, J.; Gale, M.; Ma, Y.; Yang, Y. Ellagic acid promotes browning of white adipose tissues in high-fat diet-induced obesity in rats through suppressing white adipocyte maintaining genes. Endocr. J. 2019, 66, 923–936. [Google Scholar] [CrossRef] [Green Version]
- Torriani, M.; Fitch, K.; Stavrou, E.; Bredella, M.A.; Lim, R.; Sass, C.A.; Cypess, A.M.; Grinspoon, S. Deiodinase 2 expression is increased in dorsocervical fat of patients with HIV-associated lipohypertrophy syndrome. J. Clin. Endocrinol. Metab. 2012, 97, E602–E607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minaguchi, J.A.; Ogata, S.; Takahashi, N.; Hirose, T.; Ueda, H.; Takehana, K. Remodeling of rat stromal-vascular cells to brite/beige adipocytes by prolyl-hydroxyproline. J. Vet. Med. Sci 2017, 79, 547–553. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.A.; Tao, C.; Gupta, R.K.; Scherer, P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med. 2013, 19, 1338–1344. [Google Scholar] [CrossRef]
- McGaugh, S.; Schwartz, T.S. Here and there, but not everywhere: Repeated loss of uncoupling protein 1 in amniotes. Biol. Lett. 2017, 13. [Google Scholar] [CrossRef] [Green Version]
- Van Slambrouck, S.; Jenkins, A.R.; Romero, A.E.; Steelant, W.F. Reorganization of the integrin alpha2 subunit controls cell adhesion and cancer cell invasion in prostate cancer. Int. J. Oncol. 2009, 34, 1717–1726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elices, M.J.; Hemler, M.E. The human integrin VLA-2 is a collagen receptor on some cells and a collagen/laminin receptor on others. Proc. Natl. Acad. Sci. USA 1989, 86, 9906–9910. [Google Scholar] [CrossRef] [Green Version]
- Long, J.Z.; Svensson, K.J.; Tsai, L.; Zeng, X.; Roh, H.C.; Kong, X.; Rao, R.R.; Lou, J.; Lokurkar, I.; Baur, W.; et al. A smooth muscle-like origin for beige adipocytes. Cell Metab. 2014, 19, 810–820. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Hu, S.; Cao, C.; Chen, C.; Liu, J.; Wang, Y.; Liu, J.; Zhao, J.; Tao, C.; Wang, Y. Functional and Genetic Characterization of Porcine Beige Adipocytes. Cells 2022, 11, 751. https://doi.org/10.3390/cells11040751
Zhang L, Hu S, Cao C, Chen C, Liu J, Wang Y, Liu J, Zhao J, Tao C, Wang Y. Functional and Genetic Characterization of Porcine Beige Adipocytes. Cells. 2022; 11(4):751. https://doi.org/10.3390/cells11040751
Chicago/Turabian StyleZhang, Lilan, Silu Hu, Chunwei Cao, Chuanhe Chen, Jiali Liu, Yu Wang, Jianfeng Liu, Jianguo Zhao, Cong Tao, and Yanfang Wang. 2022. "Functional and Genetic Characterization of Porcine Beige Adipocytes" Cells 11, no. 4: 751. https://doi.org/10.3390/cells11040751
APA StyleZhang, L., Hu, S., Cao, C., Chen, C., Liu, J., Wang, Y., Liu, J., Zhao, J., Tao, C., & Wang, Y. (2022). Functional and Genetic Characterization of Porcine Beige Adipocytes. Cells, 11(4), 751. https://doi.org/10.3390/cells11040751