LINC00152 Drives a Competing Endogenous RNA Network in Human Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Gene and miRNA Expression Profiling
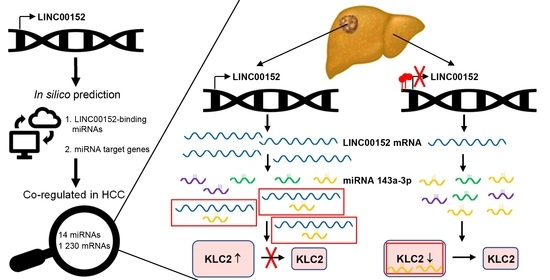
2.2. Construction and Modeling of the LINC00152-Driven ceRNA Network
2.3. RNA In Situ Hybridization Using Tissue Microarray
2.4. Cell Lines and Transfections
2.5. Functional Analyses
2.6. Total RNA, miRNA Isolation and qPCR
2.7. Western Immunoblotting
2.8. RNA Immunoprecipitation (RIP)
2.9. Statistical Analysis
3. Results
3.1. LINC00152 Upregulation Promotes Tumorigenicity of Human Liver Cancer Cells
3.2. LINC00152 Is Enriched in RNP Complexes of Potentially LINC00152 Binding miRNAs
3.3. Kinesin Light Chain 2 (KLC2) May Be a Component of a LINC00152-Driven ceRNA Network
3.4. LINC00152 Controls the Bioavailability of miR-143a-3p
3.5. KLC2 Exerts Oncogenic Functions in Human HCC
3.6. LINC00152 and KLC2 RNAs Are Co-Expressed in Human HCC Cells
3.7. KLC2 Mediates the Oncogenic Functions of the LINC00152 ceRNA Network in Liver Cancer Proliferation
4. Discussion
5. Limitations and Perspectives
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calderaro, J.; Couchy, G.; Imbeaud, S.; Amaddeo, G.; Letouze, E.; Blanc, J.F.; Laurent, C.; Hajji, Y.; Azoulay, D.; Bioulac-Sage, P.; et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J. Hepatol. 2017, 67, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Shimada, S.; Mogushi, K.; Akiyama, Y.; Furuyama, T.; Watanabe, S.; Ogura, T.; Ogawa, K.; Ono, H.; Mitsunori, Y.; Ban, D.; et al. Comprehensive molecular and immunological characterization of hepatocellular carcinoma. EBioMedicine 2019, 40, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Montironi, C.; Castet, F.; Haber, P.K.; Pinyol, R.; Torres-Martin, M.; Torrens, L.; Mesropian, A.; Wang, H.; Puigvehi, M.; Maeda, M.; et al. Inflamed and non-inflamed classes of HCC: A revised immunogenomic classification. Gut 2022, in press. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017, 169, 1327–1341. [Google Scholar] [CrossRef] [PubMed]
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- DiStefano, J.K.; Gerhard, G.S. Long noncoding RNAs and human liver disease. Annu. Rev. Pathol. 2022, 17, 1–21. [Google Scholar] [CrossRef]
- Mahpour, A.; Mullen, A.C. Our emerging understanding of the roles of long non-coding RNAs in normal liver function, disease, and malignancy. JHEP Rep. 2021, 3, 100177. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, J.K.; Peng, Y.; He, W.; Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer 2020, 19, 77. [Google Scholar] [CrossRef]
- Neumann, O.; Kesselmeier, M.; Geffers, R.; Pellegrino, R.; Radlwimmer, B.; Hoffmann, K.; Ehemann, V.; Schemmer, P.; Schirmacher, P.; Lorenzo Bermejo, J.; et al. Methylome analysis and integrative profiling of human HCCs identify novel protumorigenic factors. Hepatology 2012, 56, 1817–1827. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, J.; Li, Q.; Xu, B.; Lian, Y.; Miao, L. LINC00152: A pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017, 50, e12349. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Tang, J.; Deng, L.; Xie, Y.; Jiang, R.; Li, G.; Sun, B. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget 2015, 6, 42813–42824. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Q.; Chen, Q.; Qin, H.X.; Yu, Y.Q.; Weng, J.; Mo, Q.R.; Yin, X.F.; Lin, Y.; Liao, W.J. Long intergenic nonprotein coding RNA 0152 promotes hepatocellular carcinoma progression by regulating phosphatidylinositol 3-Kinase/Akt/Mammalian target of rapamycin signaling pathway through miR-139/PIK3CA. Am. J. Pathol. 2020, 190, 1095–1107. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Wang, H.; Sun, J.; Liu, H.; Zheng, C.; Zhou, X.; Lu, Z. LINC00152 promotes cell cycle progression in hepatocellular carcinoma via miR-193a/b-3p/CCND1 axis. Cell Cycle 2018, 17, 974–984. [Google Scholar] [CrossRef]
- Xia, T.; Liao, Q.; Jiang, X.; Shao, Y.; Xiao, B.; Xi, Y.; Guo, J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci. Rep. 2014, 4, 6088. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef]
- Longerich, T.; Breuhahn, K.; Odenthal, M.; Petmecky, K.; Schirmacher, P. Factors of transforming growth factor beta signalling are co-regulated in human hepatocellular carcinoma. Virchows Arch. 2004, 445, 589–596. [Google Scholar] [CrossRef]
- Aparicio-Prat, E.; Arnan, C.; Sala, I.; Bosch, N.; Guigo, R.; Johnson, R. DECKO: Single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genom. 2015, 16, 846. [Google Scholar] [CrossRef]
- Pulido-Quetglas, C.; Aparicio-Prat, E.; Arnan, C.; Polidori, T.; Hermoso, T.; Palumbo, E.; Ponomarenko, J.; Guigo, R.; Johnson, R. Scalable design of paired CRISPR guide RNAs for genomic deletion. PLoS Comput. Biol. 2017, 13, e1005341. [Google Scholar] [CrossRef]
- Castoldi, M.; Vujic Spasic, M.; Altamura, S.; Elmen, J.; Lindow, M.; Kiss, J.; Stolte, J.; Sparla, R.; D’Alessandro, L.A.; Klingmuller, U.; et al. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J. Clin. Investig. 2011, 121, 1386–1396. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, R.; Thavamani, A.; Calvisi, D.F.; Budczies, J.; Neumann, A.; Geffers, R.; Kroemer, J.; Greule, D.; Schirmacher, P.; Nordheim, A.; et al. Serum Response Factor (SRF) drives the transcriptional upregulation of the MDM4 oncogene in HCC. Cancers 2021, 13, 199. [Google Scholar] [CrossRef] [PubMed]
- Benes, V.; Collier, P.; Kordes, C.; Stolte, J.; Rausch, T.; Muckentaler, M.U.; Haussinger, D.; Castoldi, M. Identification of cytokine-induced modulation of microRNA expression and secretion as measured by a novel microRNA specific qPCR assay. Sci. Rep. 2015, 5, 11590. [Google Scholar] [CrossRef]
- Budczies, J.; Klauschen, F.; Sinn, B.V.; Gyorffy, B.; Schmitt, W.D.; Darb-Esfahani, S.; Denkert, C. Cutoff Finder: A comprehensive and straightforward web application enabling rapid biomarker cutoff optimization. PLoS ONE 2012, 7, e51862. [Google Scholar] [CrossRef] [PubMed]
- Izaurralde, E. Elucidating the temporal order of silencing. EMBO Rep. 2012, 13, 662–663. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, X.; Fang, D.; Xu, P.; Mo, X.; Hu, C.; Abdelatty, A.; Wang, M.; Xu, H.; Sun, Q.; et al. STK39 is a novel kinase contributing to the progression of hepatocellular carcinoma by the PLK1/ERK signaling pathway. Theranostics 2021, 11, 2108–2122. [Google Scholar] [CrossRef]
- Xu, H.; Hu, Y.W.; Zhao, J.Y.; Hu, X.M.; Li, S.F.; Wang, Y.C.; Gao, J.J.; Sha, Y.H.; Kang, C.M.; Lin, L.; et al. MicroRNA-195–5p acts as an anti-oncogene by targeting PHF19 in hepatocellular carcinoma. Oncol. Rep. 2015, 34, 175–182. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W. Long noncoding RNA CYTOR sponges miR-195 to modulate proliferation, migration, invasion and radiosensitivity in nonsmall cell lung cancer cells. Biosci. Rep. 2018, 38, BSR20181599. [Google Scholar] [CrossRef]
- Liu, Y.; Li, M.; Yu, H.; Piao, H. lncRNA CYTOR promotes tamoxifen resistance in breast cancer cells via sponging miR125a5p. Int. J. Mol. Med. 2020, 45, 497–509. [Google Scholar] [CrossRef]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Wang, B.; Yang, S.; Zhao, W. Long non-coding RNA NRAD1 and LINC00152 are highly expressed and associated with prognosis in patients with hepatocellular carcinoma. Onco. Targets Ther. 2020, 13, 10409–10416. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, X.; Tang, J.; Jiang, R.; Zhang, W.; Ji, J.; Sun, B. HULC and LINC00152 act as novel biomarkers in predicting diagnosis of hepatocellular carcinoma. Cell Physiol. Biochem. 2015, 37, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Fang, X.; Xia, B.; Zhao, Y.; Li, Q.; Wu, X. Long noncoding RNA LINC00152 promotes cell proliferation through competitively binding endogenous miR-125b with MCL-1 by regulating mitochondrial apoptosis pathways in ovarian cancer. Cancer Med. 2018, 7, 4530–4541. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Yang, X.B.; Yang, X.; Sang, X.T. LncRNA CYTOR affects the proliferation, cell cycle and apoptosis of hepatocellular carcinoma cells by regulating the miR-125b-5p/KIAA1522 axis. Aging 2020, 13, 2626–2639. [Google Scholar] [CrossRef]
- Li, X.; Rui, B.; Cao, Y.; Gong, X.; Li, H. Long non-coding RNA LINC00152 acts as a sponge of miRNA-193b-3p to promote tongue squamous cell carcinoma progression. Oncol. Lett. 2020, 19, 2035–2042. [Google Scholar] [CrossRef]
- Liu, P.; He, W.; Lu, Y.; Wang, Y. Long non-coding RNA LINC00152 promotes tumorigenesis via sponging miR-193b-3p in osteosarcoma. Oncol. Lett. 2019, 18, 3630–3636. [Google Scholar] [CrossRef]
- Wang, H.; Chen, W.; Yang, P.; Zhou, J.; Wang, K.; Tao, Q. Knockdown of LINC00152 inhibits the progression of gastric cancer by regulating microRNA-193b-3p/ETS1 axis. Cancer Biol. Ther. 2019, 20, 461–473. [Google Scholar] [CrossRef]
- Huang, Y.; Luo, H.; Li, F.; Yang, Y.; Ou, G.; Ye, X.; Li, N. LINC00152 down-regulated miR-193a-3p to enhance MCL1 expression and promote gastric cancer cells proliferation. Biosci Rep. 2018, 38, BSR20171607. [Google Scholar] [CrossRef]
- Ballare, C.; Lange, M.; Lapinaite, A.; Martin, G.M.; Morey, L.; Pascual, G.; Liefke, R.; Simon, B.; Shi, Y.; Gozani, O.; et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat. Struct. Mol. Biol. 2012, 19, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.M.; Huang, M.D.; Sun, D.P.; Kong, R.; Xu, T.P.; Xia, R.; Zhang, E.B.; Shu, Y.Q. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget 2016, 7, 9773–9787. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.N.; Chen, X.; Chen, Z.Y.; Nie, F.Q.; Wei, C.C.; Ma, H.W.; Wan, L.; Yan, S.; Ren, S.N.; Wang, Z.X. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol. Cancer 2017, 16, 17. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, R.; Calvisi, D.F.; Ladu, S.; Ehemann, V.; Staniscia, T.; Evert, M.; Dombrowski, F.; Schirmacher, P.; Longerich, T. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology 2010, 51, 857–868. [Google Scholar] [CrossRef]
- Dimri, M.; Satyanarayana, A. Molecular signaling pathways and therapeutic targets in hepatocellular carcinoma. Cancers 2020, 12, 491. [Google Scholar] [CrossRef]
- Shi, Y.; Sun, H. Down-regulation of lncRNA LINC00152 suppresses gastric cancer cell migration and invasion through inhibition of the ERK/MAPK signaling pathway. OncoTargets Ther. 2020, 13, 2115–2124. [Google Scholar] [CrossRef]
- Hirokawa, N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 1998, 279, 519–526. [Google Scholar] [CrossRef]
- Batut, J.; Howell, M.; Hill, C.S. Kinesin-mediated transport of Smad2 is required for signaling in response to TGF-beta ligands. Dev. Cell 2007, 12, 261–274. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, X.; Sha, Z.; Li, N.; Li, D.; Chen, L. High expression of kinesin light chain-2, a novel target of miR-125b, is associated with poor clinical outcome of elderly non-small-cell lung cancer patients. Br. J. Cancer 2015, 112, 874–882. [Google Scholar] [CrossRef]
- Wang, Y.L.; Liu, J.Y.; Yang, J.E.; Yu, X.M.; Chen, Z.L.; Chen, Y.J.; Kuang, M.; Zhu, Y.; Zhuang, S.M. Lnc-UCID promotes G1/S transition and hepatoma growth by preventing DHX9-mediated CDK6 down-regulation. Hepatology 2019, 70, 259–275. [Google Scholar] [CrossRef]
- Huang, M.; Wang, H.; Hu, X.; Cao, X. lncRNA MALAT1 binds chromatin remodeling subunit BRG1 to epigenetically promote inflammation-related hepatocellular carcinoma progression. Oncoimmunology 2019, 8, e1518628. [Google Scholar] [CrossRef] [PubMed]







| Gene Symbol | Correlation Value * |
|---|---|
| STK39 | 0.59425 |
| FAM60A | 0.58052 |
| FUT4 | 0.56763 |
| PALLD | 0.56334 |
| MAP3K1 | 0.55911 |
| PLAU | 0.55639 |
| C15orf39 | 0.54633 |
| ABCC5 | 0.52205 |
| PODXL | 0.52193 |
| E2F3 | 0.51697 |
| KLC2 | 0.51462 |
| LRP12 | 0.51185 |
| CHD7 | 0.50999 |
| UBE2Q2 | 0.50646 |
| PHF19 | 0.50132 |
| FAM122B | 0.50077 |
| EIF2C2 | 0.49204 |
| FAM60A | 0.48958 |
| LHFPL2 | 0.48275 |
| RAP2B | 0.47997 |
| miRNA Symbol | p Value (Adjusted) |
|---|---|
| hsa-miR-23a-3p | 0.000057 |
| hsa-miR-125a-5p | 0.000358 |
| hsa-miR-125b-5p | 0.000358 |
| hsa-miR-223-3p | 0.000401 |
| hsa-miR-143-3p | 0.001238 |
| hsa-miR-497-5p | 0.001238 |
| hsa-let-7c | 0.001870 |
| hsa-miR-150-5p | 0.002481 |
| hsa-miR-195-5p | 0.002895 |
| hsa-miR-193b-3p | 0.007043 |
| hsa.let.7b.5p | 0.011390 |
| hsa-miR.18a.5p | 0.021426 |
| hsa-miR.139.5p | 0.037874 |
| hsa-let.7a.5p | 0.049108 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pellegrino, R.; Castoldi, M.; Ticconi, F.; Skawran, B.; Budczies, J.; Rose, F.; Schwab, C.; Breuhahn, K.; Neumann, U.P.; Gaisa, N.T.; et al. LINC00152 Drives a Competing Endogenous RNA Network in Human Hepatocellular Carcinoma. Cells 2022, 11, 1528. https://doi.org/10.3390/cells11091528
Pellegrino R, Castoldi M, Ticconi F, Skawran B, Budczies J, Rose F, Schwab C, Breuhahn K, Neumann UP, Gaisa NT, et al. LINC00152 Drives a Competing Endogenous RNA Network in Human Hepatocellular Carcinoma. Cells. 2022; 11(9):1528. https://doi.org/10.3390/cells11091528
Chicago/Turabian StylePellegrino, Rossella, Mirco Castoldi, Fabio Ticconi, Britta Skawran, Jan Budczies, Fabian Rose, Constantin Schwab, Kai Breuhahn, Ulf P. Neumann, Nadine T. Gaisa, and et al. 2022. "LINC00152 Drives a Competing Endogenous RNA Network in Human Hepatocellular Carcinoma" Cells 11, no. 9: 1528. https://doi.org/10.3390/cells11091528
APA StylePellegrino, R., Castoldi, M., Ticconi, F., Skawran, B., Budczies, J., Rose, F., Schwab, C., Breuhahn, K., Neumann, U. P., Gaisa, N. T., Loosen, S. H., Luedde, T., Costa, I. G., & Longerich, T. (2022). LINC00152 Drives a Competing Endogenous RNA Network in Human Hepatocellular Carcinoma. Cells, 11(9), 1528. https://doi.org/10.3390/cells11091528