RXR Agonists Enhance Lenalidomide Anti-Myeloma Activity and T Cell Functions while Retaining Glucose-Lowering Effect
Abstract
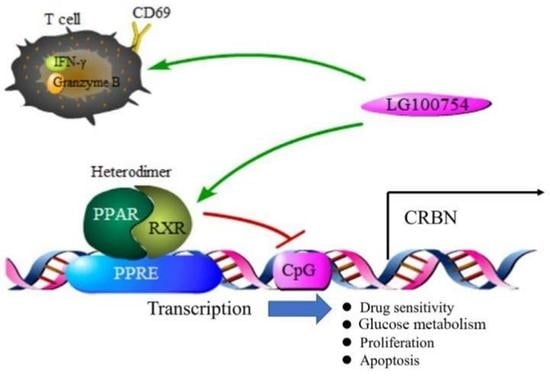
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-Associated Protein 9 (Cas9) Genomic Editing
2.3. CRBN Firefly Luciferase Reporter System
2.4. Methylation Specific PCR (MSP)
2.5. Lentiviral Gene Transduction
2.6. Co-Immunoprecipitation
2.7. Reagents and Antibodies
2.8. Thiazolyl Blue Tetrazolium Bromide (MTT) Cell Proliferation Assay
2.9. Western Blot Analysis
2.10. Flow Cytometry for the Measurement of T Cell Activation and Exhaustion
2.11. Chromatin Immunoprecipitation (ChIP) Assay
2.12. Confocal Microscopy Examination of CRBN in MM Cell Lines
2.13. In Vivo Glucose and Lipid Measurement
2.14. Myeloma Xenograft Mouse Model
2.15. Statistical Analysis
3. Results
3.1. Synergistic Anti-Myeloma Activity of Lenalidomide and RXR Agonists In Vitro
3.2. Increased CRBN Expression Correlates with IMiD and RXR Agonist Synergism
3.3. RXR Agonists Attenuate the Binding Effect of PPARs on the CRBN Promoter Region in MM Cell Lines
3.4. RXR Agonists Facilitated DNA Demethylation in the CpG Islands within the CRBN Promoter
3.5. LG100754 Improved Markers of T-Cell Activity and Suppressed T-Cell Exhaustion Markers
3.6. LG100754 Enhanced Lenalidomide’s Anti-Myeloma Activity in a Preclinical Model of MM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wenthe, J.; Naseri, S.; Hellstrom, A.C.; Wiklund, H.J.; Eriksson, E.; Loskog, A. Immunostimulatory oncolytic virotherapy for multiple myeloma targeting 4-1BB and/or CD40. Cancer Gene Ther. 2020, 27, 948–959. [Google Scholar] [CrossRef]
- van Nieuwenhuijzen, N.; Frunt, R.; May, A.M.; Minnema, M.C. Therapeutic outcome of early-phase clinical trials in multiple myeloma: A meta-analysis. Blood Cancer J. 2021, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Faruq, O.; Zhao, D.; Shrestha, M.; Vecchione, A.; Zacksenhaus, E.; Chang, H. Targeting an MDM2/MYC Axis to Overcome Drug Resistance in Multiple Myeloma. Cancers 2022, 14, 1592. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.V. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2020, 95, 548–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rieck, M.; Wedeken, L.; Muller-Brusselbach, S.; Meissner, W.; Muller, R. Expression level and agonist-binding affect the turnover, ubiquitination and complex formation of peroxisome proliferator activated receptor beta. FEBS J. 2007, 274, 5068–5076. [Google Scholar] [CrossRef]
- Wood, W.M.; Sharma, V.; Bauerle, K.T.; Pike, L.A.; Zhou, Q.; Fretwell, D.L.; Schweppe, R.E.; Haugen, B.R. PPARgamma Promotes Growth and Invasion of Thyroid Cancer Cells. PPAR Res. 2011, 2011, 171765. [Google Scholar] [CrossRef] [Green Version]
- Evans, R.M. The steroid and thyroid hormone receptor superfamily. Science 1988, 240, 889–895. [Google Scholar] [CrossRef]
- Huang, Z.; Chu, M.; Chen, X.; Wang, Z.; Jiang, L.; Ma, Y.; Wang, Y. Th2A cells: The pathogenic players in allergic diseases. Front. Immunol. 2022, 13, 916778. [Google Scholar] [CrossRef]
- Oyama, T.; Toyota, K.; Waku, T.; Hirakawa, Y.; Nagasawa, N.; Kasuga, J.I.; Hashimoto, Y.; Miyachi, H.; Morikawa, K. Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2009, 65, 786–795. [Google Scholar] [CrossRef]
- Hou, X.; Li, Y.; Huang, Y.; Zhao, H.; Gui, L. Adenosine Receptor A1-A2a Heteromers Regulate EAAT2 Expression and Glutamate Uptake via YY1-Induced Repression of PPARgamma Transcription. PPAR Res. 2020, 2020, 2410264. [Google Scholar] [CrossRef] [Green Version]
- Harmon, G.S.; Lam, M.T.; Glass, C.K. PPARs and lipid ligands in inflammation and metabolism. Chem. Rev. 2011, 111, 6321–6340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sha, Y.; Wu, J.; Paul, B.; Zhao, Y.; Mathews, P.; Li, Z.; Norris, J.; Wang, E.; McDonnell, D.P.; Kang, Y. PPAR agonists attenuate lenalidomide’s anti-myeloma activity in vitro and in vivo. Cancer Lett. 2022, 545, 215832. [Google Scholar] [CrossRef]
- Simon, D.M.; Mariani, T.J. Role of PPARs and Retinoid X Receptors in the Regulation of Lung Maturation and Development. PPAR Res. 2007, 2007, 91240. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, J.; Robertson, C.L.; Rajasekaran, D.; Gredler, R.; Siddiq, A.; Emdad, L.; Mukhopadhyay, N.D.; Ghosh, S.; Hylemon, P.B.; Gil, G.; et al. AEG-1 regulates retinoid X receptor and inhibits retinoid signaling. Cancer Res. 2014, 74, 4364–4377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, I.; Fisher, P.B.; Sarkar, D. Astrocyte elevated gene-1 (AEG-1): A key driver of hepatocellular carcinoma (HCC). Adv. Cancer Res. 2021, 152, 329–381. [Google Scholar] [CrossRef] [PubMed]
- Baba, A.; Shimizu, M.; Ohno, T.; Shirakami, Y.; Kubota, M.; Kochi, T.; Terakura, D.; Tsurumi, H.; Moriwaki, H. Synergistic growth inhibition by acyclic retinoid and phosphatidylinositol 3-kinase inhibitor in human hepatoma cells. BMC Cancer 2013, 13, 465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.C. The role of peroxisome proliferator-activated receptors in the development and physiology of gametes and preimplantation embryos. PPAR Res. 2008, 2008, 732303. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Brown, P.H. Prevention of ER-negative breast cancer. Recent Results Cancer Res. 2009, 181, 121–134. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Huang, J.; Wang, Q.; He, J.; Teng, Y.; Jiang, R.; Broxmeyer, H.E.; Guo, B. RXR Negatively Regulates Ex Vivo Expansion of Human Cord Blood Hematopoietic Stem and Progenitor Cells. Stem Cell Rev. Rep. 2021, 17, 1456–1464. [Google Scholar] [CrossRef]
- Kunej, T.; Jevsinek Skok, D.; Zorc, M.; Ogrinc, A.; Michal, J.J.; Kovac, M.; Jiang, Z. Obesity gene atlas in mammals. J. Genom. 2013, 1, 45–55. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, S.; Kitagishi, Y. Peroxisome proliferator-activated receptor and vitamin d receptor signaling pathways in cancer cells. Cancers 2013, 5, 1261–1270. [Google Scholar] [CrossRef] [Green Version]
- Nickkho-Amiry, M.; McVey, R.; Holland, C. Peroxisome proliferator-activated receptors modulate proliferation and angiogenesis in human endometrial carcinoma. Mol. Cancer Res. 2012, 10, 441–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenhard, J.M. PPAR gamma/RXR as a molecular target for diabetes. Recept. Channels 2001, 7, 249–258. [Google Scholar]
- Maimouni, S.; Issa, N.; Cheng, S.; Ouaari, C.; Cheema, A.; Kumar, D.; Byers, S. Tumor suppressor RARRES1- A novel regulator of fatty acid metabolism in epithelial cells. PLoS ONE 2018, 13, e0208756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cesario, R.M.; Klausing, K.; Razzaghi, H.; Crombie, D.; Rungta, D.; Heyman, R.A.; Lala, D.S. The rexinoid LG100754 is a novel RXR:PPARgamma agonist and decreases glucose levels in vivo. Mol. Endocrinol. 2001, 15, 1360–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.; Ramalanjaona, N.; Huet, T.; Potier, N.; Osz, J.; Antony, P.; Peluso-Iltis, C.; Poussin-Courmontagne, P.; Ennifar, E.; Mely, Y.; et al. The "Phantom Effect" of the Rexinoid LG100754: Structural and functional insights. PLoS ONE 2010, 5, e15119. [Google Scholar] [CrossRef] [Green Version]
- Forman, B.M. The antidiabetic agent LG100754 sensitizes cells to low concentrations of peroxisome proliferator-activated receptor gamma ligands. J. Biol. Chem. 2002, 277, 12503–12506. [Google Scholar] [CrossRef] [Green Version]
- Shen, D.; Yu, X.; Wu, Y.; Chen, Y.; Li, G.; Cheng, F.; Xia, L. Emerging roles of bexarotene in the prevention, treatment and anti-drug resistance of cancers. Expert Rev. Anticancer Ther. 2018, 18, 487–499. [Google Scholar] [CrossRef]
- He, J.; Huang, Y.; Liu, H.; Sun, X.; Wu, J.; Zhang, Z.; Liu, L.; Zhou, C.; Jiang, S.; Huang, Z.; et al. Bexarotene promotes microglia/macrophages—Specific brain—Derived Neurotrophic factor expression and axon sprouting after traumatic brain injury. Exp. Neurol. 2020, 334, 113462. [Google Scholar] [CrossRef]
- Saito-Hakoda, A.; Uruno, A.; Yokoyama, A.; Shimizu, K.; Parvin, R.; Kudo, M.; Saito-Ito, T.; Sato, I.; Kogure, N.; Suzuki, D.; et al. Effects of RXR Agonists on Cell Proliferation/Apoptosis and ACTH Secretion/Pomc Expression. PLoS ONE 2015, 10, e0141960. [Google Scholar] [CrossRef] [Green Version]
- Scheepstra, M.; Andrei, S.A.; de Vries, R.; Meijer, F.A.; Ma, J.N.; Burstein, E.S.; Olsson, R.; Ottmann, C.; Milroy, L.G.; Brunsveld, L. Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization. ACS Chem. Neurosci. 2017, 8, 2065–2077. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, S.; Taniguchi, H.; Moritoh, Y.; Tashiro, F.; Yamamoto, T.; Yamato, E.; Ikegami, H.; Ozato, K.; Miyazaki, J. Nuclear hormone retinoid X receptor (RXR) negatively regulates the glucose-stimulated insulin secretion of pancreatic ss-cells. Diabetes 2010, 59, 2854–2861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stathopoulou, A.; Chhetri, J.B.; Ambrose, J.C.; Esteve, P.O.; Ji, L.; Erdjument-Bromage, H.; Zhang, G.; Neubert, T.A.; Pradhan, S.; Herrero, J.; et al. A novel requirement for DROSHA in maintenance of mammalian CG methylation. Nucleic Acids Res. 2017, 45, 9398–9412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, D.; Van Meir, E.G. A simple genotyping method to detect small CRISPR-Cas9 induced indels by agarose gel electrophoresis. Sci. Rep. 2019, 9, 4437. [Google Scholar] [CrossRef] [Green Version]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Manosalva Perez, N.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Misawa, K.; Ueda, Y.; Kanazawa, T.; Misawa, Y.; Jang, I.; Brenner, J.C.; Ogawa, T.; Takebayashi, S.; Grenman, R.A.; Herman, J.G.; et al. Epigenetic inactivation of galanin receptor 1 in head and neck cancer. Clin. Cancer Res. 2008, 14, 7604–7613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, J.H.; Lee, B.H.; Kim, S.E.; Kwon, B.E.; Jeong, H.; Choi, J.R.; Kim, M.J.; Park, Y.; Kim, B.S.; Kim, D.H.; et al. A Novel Anti-PD-L1 Antibody Exhibits Antitumor Effects on Multiple Myeloma in Murine Models via Antibody-Dependent Cellular Cytotoxicity. Biomol. Ther. 2021, 29, 166–174. [Google Scholar] [CrossRef]
- Huh, H.D.; Sub, Y.; Oh, J.; Kim, Y.E.; Lee, J.Y.; Kim, H.R.; Lee, S.; Lee, H.; Pak, S.; Amos, S.E.; et al. Reprogramming anchorage dependency by adherent-to-suspension transition promotes metastatic dissemination. Mol. Cancer 2023, 22, 63. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, M.; Faruq, O.; Zacksenhaus, E.; Chen, W.; Liu, A.; Chang, H. SMAD1 as a biomarker and potential therapeutic target in drug-resistant multiple myeloma. Biomark Res. 2021, 9, 48. [Google Scholar] [CrossRef]
- Ding, B.; Wahid, M.A.; Wang, Z.; Xie, C.; Thakkar, A.; Prabhu, S.; Wang, J. Triptolide and celastrol loaded silk fibroin nanoparticles show synergistic effect against human pancreatic cancer cells. Nanoscale 2017, 9, 11739–11753. [Google Scholar] [CrossRef]
- Yang, T.; Liu, X.; Kumar, S.K.; Jin, F.; Dai, Y. Decoding DNA methylation in epigenetics of multiple myeloma. Blood Rev. 2022, 51, 100872. [Google Scholar] [CrossRef]
- Muylaert, C.; Van Hemelrijck, L.A.; Maes, A.; De Veirman, K.; Menu, E.; Vanderkerken, K.; De Bruyne, E. Aberrant DNA methylation in multiple myeloma: A major obstacle or an opportunity? Front. Oncol. 2022, 12, 979569. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chu, E.; Paul, B.; Kang, Y. Mechanistic Studies and a Retrospective Cohort Study: The Interaction between PPAR Agonists and Immunomodulatory Agents in Multiple Myeloma. Cancers 2022, 14, 5272. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yu, C.; Zhuang, S. Histone Methyltransferase EZH2: A Potential Therapeutic Target for Kidney Diseases. Front. Physiol. 2021, 12, 640700. [Google Scholar] [CrossRef] [PubMed]
- Ushio, R.; Hiroi, M.; Matsumoto, A.; Mori, K.; Yamamoto, N.; Ohmori, Y. Enhanced Cytotoxic Effects in Human Oral Squamous Cell Carcinoma Cells Treated with Combined Methyltransferase Inhibitors and Histone Deacetylase Inhibitors. Biomedicines 2022, 10, 763. [Google Scholar] [CrossRef]
- Zhao, R.; Pan, Z.; Li, B.; Zhao, S.; Zhang, S.; Qi, Y.; Qiu, J.; Gao, Z.; Fan, Y.; Guo, Q.; et al. Comprehensive Analysis of the Tumor Immune Microenvironment Landscape in Glioblastoma Reveals Tumor Heterogeneity and Implications for Prognosis and Immunotherapy. Front. Immunol. 2022, 13, 820673. [Google Scholar] [CrossRef]
- Chalmin, F.; Bruchard, M.; Vegran, F.; Ghiringhelli, F. Regulation of T cell antitumor immune response by tumor induced metabolic stress. Cell Stress 2018, 3, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Kawano, Y.; Moschetta, M.; Manier, S.; Glavey, S.; Gorgun, G.T.; Roccaro, A.M.; Anderson, K.C.; Ghobrial, I.M. Targeting the bone marrow microenvironment in multiple myeloma. Immunol. Rev. 2015, 263, 160–172. [Google Scholar] [CrossRef]
- Emmons, T.R.; Giridharan, T.; Singel, K.L.; Khan, A.N.H.; Ricciuti, J.; Howard, K.; Silva-Del Toro, S.L.; Debreceni, I.L.; Aarts, C.E.M.; Brouwer, M.C.; et al. Mechanisms Driving Neutrophil-Induced T-cell Immunoparalysis in Ovarian Cancer. Cancer Immunol. Res. 2021, 9, 790–810. [Google Scholar] [CrossRef]
- Chang, C.H.; Qiu, J.; O’Sullivan, D.; Buck, M.D.; Noguchi, T.; Curtis, J.D.; Chen, Q.; Gindin, M.; Gubin, M.M.; van der Windt, G.J.; et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 2015, 162, 1229–1241. [Google Scholar] [CrossRef] [Green Version]
- Barnaba, V. T Cell Memory in Infection, Cancer, and Autoimmunity. Front. Immunol. 2021, 12, 811968. [Google Scholar] [CrossRef]
- Gandhi, A.K.; Kang, J.; Havens, C.G.; Conklin, T.; Ning, Y.; Wu, L.; Ito, T.; Ando, H.; Waldman, M.F.; Thakurta, A.; et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.). Br. J. Haematol. 2014, 164, 811–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szudy-Szczyrek, A.; Mlak, R.; Szczyrek, M.; Chocholska, S.; Sompor, J.; Nogalski, A.; Malecka-Massalska, T.; Hus, M. Polymorphisms in the promoter region of the CRBN gene as a predictive factor for the first-line CTD therapy in multiple myeloma patients. Oncotarget 2018, 9, 24054–24068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barankiewicz, J.; Salomon-Perzynski, A.; Misiewicz-Krzeminska, I.; Lech-Maranda, E. CRL4(CRBN) E3 Ligase Complex as a Therapeutic Target in Multiple Myeloma. Cancers 2022, 14, 4492. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.D.; Correa, M.; Nagy, M.A.; Alexander, M.; Plantevin, V.; Grant, V.; Whitefield, B.; Huang, D.; Kercher, T.; Harris, R.; et al. Discovery of CRBN E3 Ligase Modulator CC-92480 for the Treatment of Relapsed and Refractory Multiple Myeloma. J. Med. Chem. 2020, 63, 6648–6676. [Google Scholar] [CrossRef]
- Hidalgo, J.A.; Florez, A.; Agurto, C.; Pinedo, Y.; Ayarza, R.; Rodriguez, L.; La Rosa, A.; Gutierrez, R. Metabolic and Cardiovascular Comorbidities Among Clinically Stable HIV Patients on Long-Term ARV Therapy in Five Ambulatory Clinics in Lima-Callao, Peru. Open AIDS J. 2018, 12, 126–135. [Google Scholar] [CrossRef]
- Sehgal, K.; Fadel, H.J.; Tande, A.J.; Pardi, D.S.; Khanna, S. Outcomes in Patients with SARS-CoV-2 and Clostridioides difficile Coinfection. Infect. Drug Resist. 2021, 14, 1645–1648. [Google Scholar] [CrossRef]
- Hsu, A.K.; Quach, H.; Tai, T.; Prince, H.M.; Harrison, S.J.; Trapani, J.A.; Smyth, M.J.; Neeson, P.; Ritchie, D.S. The immunostimulatory effect of lenalidomide on NK-cell function is profoundly inhibited by concurrent dexamethasone therapy. Blood 2011, 117, 1605–1613. [Google Scholar] [CrossRef] [Green Version]
- Elsakka, A.M.A.; Bary, M.A.; Abdelzaher, E.; Elnaggar, M.; Kalamian, M.; Mukherjee, P.; Seyfried, T.N. Management of Glioblastoma Multiforme in a Patient Treated With Ketogenic Metabolic Therapy and Modified Standard of Care: A 24-Month Follow-Up. Front. Nutr. 2018, 5, 20. [Google Scholar] [CrossRef] [Green Version]
- Chandra, V.; Huang, P.; Hamuro, Y.; Raghuram, S.; Wang, Y.; Burris, T.P.; Rastinejad, F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature 2008, 456, 350–356. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Kondal, M.D.; Ahmad, A.; Zhu, R.; Fan, L.; Zaborniak, P.; Madden, K.S.; de Souza, J.V.; Bronowska, A.K. Mechanistic Investigation of the Androgen Receptor DNA-Binding Domain and Modulation via Direct Interactions with DNA Abasic Sites: Understanding the Mechanisms Involved in Castration-Resistant Prostate Cancer. Int. J. Mol. Sci. 2023, 24, 1270. [Google Scholar] [CrossRef]
- Gong, X.; Marisiddaiah, R.; Rubin, L.P. Inhibition of pulmonary beta-carotene 15, 15’-oxygenase expression by glucocorticoid involves PPARalpha. PLoS ONE 2017, 12, e0181466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, L.S.; Wells, R.A. Cross-Talk between PPARs and the Partners of RXR: A Molecular Perspective. PPAR Res. 2009, 2009, 925309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, T.; Shan, S.; Li, Z.B.; Wu, Z.W.; Liao, C.Z.; Ko, B.; Lu, X.P.; Cheng, J.; Ning, Z.Q. A new retinoid-like compound that activates peroxisome proliferator-activated receptors and lowers blood glucose in diabetic mice. Biol. Pharm. Bull. 2005, 28, 1192–1196. [Google Scholar] [CrossRef] [Green Version]
- Gaunt, C.M.; Rainbow, D.B.; Mackenzie, R.J.; Jarvis, L.B.; Mousa, H.S.; Cunniffe, N.; Georgieva, Z.; Brown, J.W.; Coles, A.J.; Jones, J.L. The MS Remyelinating Drug Bexarotene (an RXR Agonist) Promotes Induction of Human Tregs and Suppresses Th17 Differentiation In Vitro. Front. Immunol. 2021, 12, 712241. [Google Scholar] [CrossRef]
- Tachibana, M.; Shinohara, M.; Yamazaki, Y.; Liu, C.C.; Rogers, J.; Bu, G.; Kanekiyo, T. Rescuing effects of RXR agonist bexarotene on aging-related synapse loss depend on neuronal LRP1. Exp. Neurol. 2016, 277, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewandowski, C.T.; Laham, M.S.; Thatcher, G.R.J. Remembering your A, B, C’s: Alzheimer’s disease and ABCA1. Acta Pharm. Sin B 2022, 12, 995–1018. [Google Scholar] [CrossRef] [PubMed]
- Leibowitz, M.D.; Ardecky, R.J.; Boehm, M.F.; Broderick, C.L.; Carfagna, M.A.; Crombie, D.L.; D’Arrigo, J.; Etgen, G.J.; Faul, M.M.; Grese, T.A.; et al. Biological characterization of a heterodimer-selective retinoid X receptor modulator: Potential benefits for the treatment of type 2 diabetes. Endocrinology 2006, 147, 1044–1053. [Google Scholar] [CrossRef]
- van Neerven, S.; Kampmann, E.; Mey, J. RAR/RXR and PPAR/RXR signaling in neurological and psychiatric diseases. Prog. Neurobiol. 2008, 85, 433–451. [Google Scholar] [CrossRef]
- Komar, C.M. Peroxisome proliferator-activated receptors (PPARs) and ovarian function—Implications for regulating steroidogenesis, differentiation, and tissue remodeling. Reprod. Biol. Endocrinol. 2005, 3, 41. [Google Scholar] [CrossRef] [Green Version]
- Peng, L.; Yang, H.; Ye, Y.; Ma, Z.; Kuhn, C.; Rahmeh, M.; Mahner, S.; Makrigiannakis, A.; Jeschke, U.; von Schonfeldt, V. Role of Peroxisome Proliferator-Activated Receptors (PPARs) in Trophoblast Functions. Int. J. Mol. Sci. 2021, 22, 433. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Wan, Y. Thiazolidinediones on PPARgamma: The Roles in Bone Remodeling. PPAR Res. 2011, 2011, 867180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Lee, N.; Park, E.S.; Yun, H.; Ha, T.U.; Jeon, H.; Yu, J.; Choi, S.; Shin, B.; Yu, J.; et al. T-Cell Death Associated Gene 51 Is a Novel Negative Regulator of PPARgamma That Inhibits PPARgamma-RXRalpha Heterodimer Formation in Adipogenesis. Mol. Cells 2021, 44, 1–12. [Google Scholar] [CrossRef]
- Leal, A.S.; Zydeck, K.; Carapellucci, S.; Reich, L.A.; Zhang, D.; Moerland, J.A.; Sporn, M.B.; Liby, K.T. Retinoid X receptor agonist LG100268 modulates the immune microenvironment in preclinical breast cancer models. NPJ Breast Cancer 2019, 5, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leal, A.S.; Moerland, J.A.; Zhang, D.; Carapellucci, S.; Lockwood, B.; Krieger-Burke, T.; Aleiwi, B.; Ellsworth, E.; Liby, K.T. The RXR Agonist MSU42011 Is Effective for the Treatment of Preclinical HER2+ Breast Cancer and Kras-Driven Lung Cancer. Cancers 2021, 13, 5004. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Wang, X.; Zhang, M.; Mathews, P.; Kang, Y. RXR Agonists Enhance Lenalidomide Anti-Myeloma Activity and T Cell Functions while Retaining Glucose-Lowering Effect. Cells 2023, 12, 1993. https://doi.org/10.3390/cells12151993
Wu J, Wang X, Zhang M, Mathews P, Kang Y. RXR Agonists Enhance Lenalidomide Anti-Myeloma Activity and T Cell Functions while Retaining Glucose-Lowering Effect. Cells. 2023; 12(15):1993. https://doi.org/10.3390/cells12151993
Chicago/Turabian StyleWu, Jian, Xiaobei Wang, Min Zhang, Parker Mathews, and Yubin Kang. 2023. "RXR Agonists Enhance Lenalidomide Anti-Myeloma Activity and T Cell Functions while Retaining Glucose-Lowering Effect" Cells 12, no. 15: 1993. https://doi.org/10.3390/cells12151993
APA StyleWu, J., Wang, X., Zhang, M., Mathews, P., & Kang, Y. (2023). RXR Agonists Enhance Lenalidomide Anti-Myeloma Activity and T Cell Functions while Retaining Glucose-Lowering Effect. Cells, 12(15), 1993. https://doi.org/10.3390/cells12151993