Comparative Shotgun Proteomics Reveals the Characteristic Protein Signature of Osteosarcoma Subtypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients Details
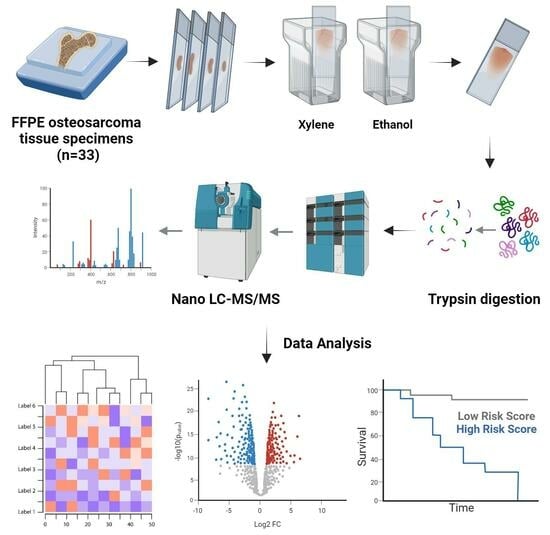
2.2. Tissue Sample Processing
2.3. LC–MS/MS Analysis
2.4. Clinical Data Analysis
2.5. Proteomics Identification
2.6. Proteomic Data Analysis
3. Results
3.1. Patient Cohort Description, Clinical Characteristics, and Outcomes
3.2. Clinical Determinants Do Not Correlate with the Proteome Signature of Pediatric Osteosarcoma
3.3. Pathological Subtypes Are Defined by Different Proteome Signatures
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ottaviani, G.; Jaffe, N. The Epidemiology of Osteosarcoma. Cancer Treat Res. 2009, 152, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Corre, I.; Verrecchia, F.; Crenn, V.; Redini, F.; Trichet, V. The Osteosarcoma Microenvironment: A Complex but Targetable Ecosystem. Cells 2020, 9, 976. [Google Scholar] [CrossRef] [PubMed]
- Isakoff, M.S.; Bielack, S.S.; Meltzer, P.; Gorlick, R. Osteosarcoma: Current Treatment and a Collaborative Pathway to Success. J. Clin. Oncol. 2015, 33, 3029–3035. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.A.; Hawkins, C.J. Recent and Ongoing Research into Metastatic Osteosarcoma Treatments. Int. J. Mol. Sci. 2022, 23, 3817. [Google Scholar] [CrossRef]
- Wadhwa, N. Osteosarcoma: Diagnostic dilemmas in histopathology and prognostic factors. Indian J. Orthop. 2014, 48, 247–254. [Google Scholar] [CrossRef]
- Yoshida, A. Osteosarcoma: Old and New Challenges. Surg. Pathol. Clin. 2021, 14, 567–583. [Google Scholar] [CrossRef] [PubMed]
- Morrow, J.J.; Khanna, C. Osteosarcoma Genetics and Epigenetics: Emerging Cancer Treat ResBiology and Candidate Therapies. Crit. Rev. Oncog. 2015, 20, 173–197. [Google Scholar] [CrossRef] [PubMed]
- Hameed, M.; Mandelker, D. Tumor Syndromes Predisposing to Osteosarcoma. Adv. Anat. Pathol. 2018, 25, 217–222. [Google Scholar] [CrossRef]
- Bernardini, G.; Laschi, M.; Geminiani, M.; Santucci, A. Proteomics of osteosarcoma. Expert Rev. Proteom. 2014, 11, 331–343. [Google Scholar] [CrossRef]
- Matos, L.L.; Trufelli, D.C.; de Matos, M.G.L.; da Silva Pinhal, M.A. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark Insights 2010, 5, 9–20. [Google Scholar] [CrossRef]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic Quotient Normalization as Robust Method to Account for Dilution of Complex Biological Mixtures. Application in 1H NMR Metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef]
- Overmyer, K.A.; Shishkova, E.; Miller, I.J.; Balnis, J.; Bernstein, M.N.; Peters-Clarke, T.M.; Meyer, J.G.; Quan, Q.; Muehlbauer, L.K.; Trujillo, E.A.; et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2020, 12, 23–40.e7. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-Protein Interaction Networks, Integrated Over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Misaghi, A.; Goldin, A.; Awad, M.; Kulidjian, A.A. Osteosarcoma: A comprehensive review. SICOT-J 2018, 4, 12. [Google Scholar] [CrossRef]
- Lindsey, B.A.; Markel, J.E.; Kleinerman, E.S. Osteosarcoma Overview. Rheumatol. Ther. 2016, 4, 25–43. [Google Scholar] [CrossRef]
- Sumithra, B.; Saxena, U.; Das, A.B. A comprehensive study on genome-wide coexpression network of KHDRBS1/Sam68 reveals its cancer and patient-specific association. Sci. Rep. 2019, 9, 11083. [Google Scholar] [CrossRef]
- Elman, J.S.; Ni, T.K.; Mengwasser, K.E.; Jin, D.; Wronski, A.; Elledge, S.J.; Kuperwasser, C. Identification of FUBP1 as a Long Tail Cancer Driver and Widespread Regulator of Tumor Suppressor and Oncogene Alternative Splicing. Cell Rep. 2019, 28, 3435–3449.e5. [Google Scholar] [CrossRef]
- Maroni, P.; Luzzati, A.; Perrucchini, G.; Cannavò, L.; Bendinelli, P. Leptin, Leptin Receptor, KHDRBS1 (KH RNA Binding Domain Containing, Signal Transduction Associated 1), and Adiponectin in Bone Metastasis from Breast Carcinoma: An Immunohistochemical Study. Biomedicines 2020, 8, 510. [Google Scholar] [CrossRef]
- Chen, S.; Li, Y.; Zhi, S.; Ding, Z.; Wang, W.; Peng, Y.; Huang, Y.; Zheng, R.; Yu, H.; Wang, J.; et al. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m6A-dependent manner. Cell Death Dis. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Liu, H.; Qin, G.; Ji, Y.; Wang, X.; Bao, H.; Guan, X.; Wei, A.; Cai, Z. Potential role of m6A RNA methylation regulators in osteosarcoma and its clinical prognostic value. J. Orthop. Surg. Res. 2021, 16, 104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Ying, L.; Tao, S.; Shi, M.; Lin, P.; Wang, Y.; Han, B. Regulatory Role of N6-methyladenosine (m6A) Modification in Osteosarcoma. Front. Oncol. 2021, 11, 683768. [Google Scholar] [CrossRef]
- Han, J.; Kong, H.; Wang, X.; Zhang, X. Novel insights into the interaction between N6-methyladenosine methylation and noncoding RNAs in muscu-loskeletal disorders. Cell Prolif. 2022, 55, e13294. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 2019, 18, 176. [Google Scholar] [CrossRef]
- Chen, X.; Xu, M.; Xu, X.; Zeng, K.; Liu, X.; Pan, B.; Li, C.; Sun, L.; Qin, J.; Xu, T.; et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol. Cancer 2020, 19, 106. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.-J.; Deng, W.-Q.; Fan, S.-R.; Chen, M.-F.; Qi, M.; Lyu, W.-Y.; Qi, Q.; Tiwari, A.K.; Chen, J.-X.; Zhang, D.-M.; et al. m6A modification: Recent advances, anticancer targeted drug discovery and beyond. Mol. Cancer 2022, 21, 52. [Google Scholar] [CrossRef]
- Sangle, N.A.; Layfield, L.J. Telangiectatic osteosarcoma. Arch. Pathol. Lab. Med. 2012, 136, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, D.J.; Nightingale, T.D.; Zenner, H.L.; Lui-Roberts, W.W.; Cutler, D.F. Formation and function of Weibel-Palade bodies. J. Cell Sci. 2008, 121, 19–27. [Google Scholar] [CrossRef]
- Hu, F.; Shang, X.-F.; Wang, W.; Jiang, W.; Fang, C.; Tan, D.; Zhou, H.-C. High-level expression of periostin is significantly correlated with tumour angiogenesis and poor prognosis in osteosarcoma. Int. J. Exp. Pathol. 2016, 97, 86–92. [Google Scholar] [CrossRef]
- Chaiyawat, P.; Sungngam, P.; Teeyakasem, P.; Sirikaew, N.; Klangjorhor, J.; Settakorn, J.; Diskul-Na-Ayudthaya, P.; Chokchaichamnankit, D.; Srisomsap, C.; Svasti, J.; et al. Protein profiling of osteosarcoma tissue and soft callus unveils activation of the unfolded protein response pathway. Int. J. Oncol. 2019, 54, 1704–1718. [Google Scholar] [CrossRef]
- Bateman, L.A.; Nguyen, T.B.; Roberts, A.M.; Miyamoto, D.K.; Ku, W.-M.; Huffman, T.R.; Petri, Y.; Heslin, M.J.; Contreras, C.M.; Skibola, C.F.; et al. Chemoproteomics-enabled covalent ligand screen reveals a cysteine hotspot in reticulon 4 that impairs ER morphology and cancer pathogenicity. Chem. Commun. 2017, 53, 7234–7237. [Google Scholar] [CrossRef]
- Chen, C.-C.; Yang, J.-H.; Fu, S.-L.; Lin, W.-J.; Lin, C.-H. Arginine Methylation of hnRNPK Inhibits the DDX3-hnRNPK Interaction to Play an Anti-Apoptosis Role in Osteosarcoma Cells. Int. J. Mol. Sci. 2021, 22, 9764. [Google Scholar] [CrossRef]
- Wang, L.; Xue, G.-B. Catalpol suppresses osteosarcoma cell proliferation through blocking epithelial-mesenchymal transition (EMT) and inducing apoptosis. Biochem. Biophys. Res. Commun. 2017, 495, 27–34. [Google Scholar] [CrossRef]
- Lei, Y.; Junxin, C.; Yongcan, H.; Xiaoguang, L.; Binsheng, Y. Role of microRNAs in the crosstalk between osteosarcoma cells and the tumour microenvironment. J. Bone Oncol. 2020, 25, 100322. [Google Scholar] [CrossRef]
- Yang, Z.; Li, X.; Yang, Y.; He, Z.; Qu, X.; Zhang, Y. Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death Dis. 2016, 7, e2389. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Xu, D.; Chen, X.; Li, S.; Zhang, L.; Chan, M.T.V.; Wu, W.K.K. An update on the roles of circular RNAs in osteosarcoma. Cell Prolif. 2020, 54, e12936. [Google Scholar] [CrossRef]
- Jolly, M.K.; Ware, K.E.; Xu, S.; Gilja, S.; Shetler, S.; Yang, Y.; Wang, X.; Austin, R.G.; Runyambo, D.; Hish, A.J.; et al. E-Cadherin Represses Anchorage-Independent Growth in Sarcomas through Both Signaling and Mechanical Mechanisms. Mol. Cancer Res. 2019, 17, 1391–1402. [Google Scholar] [CrossRef] [PubMed]
- Kashima, T.; Kawaguchi, J.; Takeshita, S.; Kuroda, M.; Takanashi, M.; Horiuchi, H.; Imamura, T.; Ishikawa, Y.; Ishida, T.; Mori, S.; et al. Anomalous Cadherin Expression in Osteosarcoma: Possible Relationships to Metastasis and Morphogenesis. Am. J. Pathol. 1999, 155, 1549–1555. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Liu, J.; Shen, G. Low expression of IGFBP4 and TAGLN accelerate the poor overall survival of osteosarcoma. Sci. Rep. 2022, 12, 9298. [Google Scholar] [CrossRef] [PubMed]
- Nikitovic, D.; Berdiaki, A.; Zafiropoulos, A.; Katonis, P.; Tsatsakis, A.; Karamanos, N.K.; Tzanakakis, G.N. Lumican expression is positively correlated with the differentiation and negatively with the growth of human osteosarcoma cells. FEBS J. 2007, 275, 350–361. [Google Scholar] [CrossRef]
- Nikitovic, D.; Chalkiadaki, G.; Berdiaki, A.; Aggelidakis, J.; Katonis, P.; Karamanos, N.K.; Tzanakakis, G.N. Lumican regulates osteosarcoma cell adhesion by modulating TGFbeta2 activity. Int. J. Biochem. Cell Biol. 2011, 43, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Heng, L.; Jia, Z.; Bai, J.; Zhang, K.; Zhu, Y.; Ma, J.; Zhang, J.; Duan, H. Molecular characterization of metastatic osteosarcoma: Differentially expressed genes, transcription factors and microRNAs. Mol. Med. Rep. 2017, 15, 2829–2836. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.-L.; Chen, G.; Song, Z.-L.; Sun, J.; Gao, X.-H.; Han, Y.-X. COL6A3 promotes cellular malignancy of osteosarcoma by activating the PI3K/AKT pathway. Rev. Assoc. Médica Bras. 2020, 66, 740–745. [Google Scholar] [CrossRef]
- Ungefroren, H.; Cikós, T.; Krull, N.B.; Kalthoff, H. Biglycan Gene Promoter Activity in Osteosarcoma Cells Is Regulated by Cyclic AMP. Biochem. Biophys. Res. Commun. 1997, 235, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Wang, B.; Qu, X.-L.; Zheng, B.-Q.; Huang, W.-D.; Sun, Z.-W.; Wang, C.-M.; Chen, Y. Metabolism of Reactive Oxygen Species in Osteosarcoma and Potential Treatment Applications. Cells 2019, 9, 87. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Liu, Y.; Han, Z.; Tian, R. Identification of potential gene signatures associated with osteosarcoma by integrated bioinformatics analysis. PeerJ 2021, 9, e11496. [Google Scholar] [CrossRef]
- Yao, F.; Zhu, Z.F.; Wen, J.; Zhang, F.Y.; Zhang, Z.; Zhu, L.Q.; Su, G.H.; Yuan, Q.W.; Zhen, Y.F.; Wang, X.D. PODN is a prognostic biomarker and correlated with immune infiltrates in osteosarcoma. Cancer Cell Int. 2021, 21, 245. [Google Scholar] [CrossRef]
- Lilienthal, I.; Herold, N. Targeting Molecular Mechanisms Underlying Treatment Efficacy and Resistance in Osteosarcoma: A Review of Current and Future Strategies. Int. J. Mol. Sci. 2020, 21, 6885. [Google Scholar] [CrossRef]
- Valdés, A.; Bitzios, A.; Kassa, E.; Shevchenko, G.; Falk, A.; Malmström, P.-U.; Dragomir, A.; Segersten, U.; Lind, S.B. Proteomic comparison between different tissue preservation methods for identification of promising biomarkers of urothelial bladder cancer. Sci. Rep. 2021, 11, 7595. [Google Scholar] [CrossRef]
- Davalieva, K.; Kiprijanovska, S.; Dimovski, A.; Rosoklija, G.; Dwork, A.J. Comparative evaluation of two methods for LC-MS/MS proteomic analysis of formalin fixed and paraffin embedded tissues. J. Proteom. 2021, 235, 104117. [Google Scholar] [CrossRef]



| Variable | No. of Cases |
|---|---|
| Age | 33 |
| Mean =12.85 | |
| Median =13.32 | |
| Age group | |
| <10 | 9 |
| ≥10 | 24 |
| Gender | 33 |
| Female | 17 |
| Male | 16 |
| Pathology | 33 |
| Conventional Subgroup | 28 |
| Chondroblastic | 9/28 |
| Fibroblastic | 5/28 |
| Osteoblastic | 14/28 |
| Non-conventional Subgroup | 5 |
| Telangectatic | 5/5 |
| Histological response to chemotherapy | 30 |
| Good (≥90% necrosis) | 7 |
| Bad (<90% necrosis) | 23 |
| Initial metastasis | 33 |
| Yes | 10 |
| No | 23 |
| Type of event | 33 |
| Event | 21 |
| >1 year-to-event | 8/21 |
| ≤1 year-to-event | 13/21 |
| No event | 12 |
| Site of event | 21 |
| Local recurrence | 1 |
| Local recurrence and Distant Metastasis | 4 |
| Distant Metastasis | 16 |
| Relative tumor size * | 31 |
| Mean = 629.2 | |
| Median = 268.9 | |
| Tumor volume | 31 |
| ≥200 | 19 |
| <200 | 12 |
| Variable | Number of Cases | OS | EFS | ||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | ||
| Pathology | 30 | 0.5 | 0.7 | ||||
| Conventional vs. Non-conventional | 25 vs. 5 | 1.741 | 0.1319–2.502 | 0.46 | 0.8034 | 0.2342–2.756 | 0.728 |
| Conventional | 28 | 0.8 | |||||
| Fibroblastic vs. Chondroblastic | 4 vs. 8 | 1.6688 | 0.3223–8.641 | 0.542 | 0.8729 | 0.2074–3.674 | 0.853 |
| Fibroblastic vs. Osteoblastic | 4 vs. 13 | 1.7914 | 0.3843–8.350 | 0.458 | 1.0878 | 0.2926–4.044 | 0.900 |
| Histological response to chemotherapy | 30 | 0.01 * | 0.005 * | ||||
| Good (≥90% necrosis) vs. Bad (<90% necrosis) | 7 vs. 23 | 0.121 | 0.01602–0.9132 | 0.0406 * | 0.9656 | 0.01282–0.7273 | 0.0233 * |
| Age group | 30 | 0.06 | 0.09 | ||||
| <10 vs. ≥10 | 8 vs. 22 | 0.4068 | 0.1523–1.087 | 0.0729 | 0.454 | 0.1784–1.156 | 0.0976 |
| Gender | 30 | 0.4 | 0.3 | ||||
| Female vs. Male | 15 vs. 15 | 1.48 | 0.5825–3.759 | 0.41 | 1.563 | 0.6452–3.778 | 0.322 |
| Initial metastasis | 30 | 0.2 | 0.09 | ||||
| Yes vs. No | 9 vs. 21 | 1.795 | 0.6947–4.636 | 0.227 | 2.177 | 0.877–5.402 | 0.0936 |
| Tumor Volume | 28 | 0.9 | 0.6 | ||||
| ≥200 vs. <200 | 18 vs. 10 | 1.038 | 0.3763–2.866 | 0.942 | 0.7848 | 0.4774–3.401 | 0.629 |
| Years-to-event | 20 | 0.00003 **** | Not Applicable | ||||
| >1 vs. ≤1 | 8 vs. 12 | 0.0365 | 0.00445–0.2993 | 0.00205 **** | |||
| Variable | OS | EFS | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| Histological response to chemotherapy | 0.1137 | 0.0150–0.8618 | 0.0354 * | 0.1051 | 0.01382–0.7989 | 0.0295 * |
| Age group | 0.9066 | 0.8014–1.0256 | 0.1192 | 0.9046 | 0.79947–1.0236 | 0.1118 |
| Initial metastasis | Not Applicable | 1.8208 | 0.70743–4.6862 | 0.2141 | ||
| Variable | Number of Cases | OS | ||
|---|---|---|---|---|
| HR | 95% CI | p-Value | ||
| Histological response to chemotherapy | 20 | 0.49406 | 0.056282–4.337 | 0.52466 |
| Age group | 20 | 1.04510 | 0.908952–1.202 | 0.53564 |
| Years-to-event | 20 | 0.02558 | 0.002617–0.250 | 0.00162 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alaa, M.; Al-Shehaby, N.; Anwar, A.M.; Farid, N.; Shawky, M.S.; Zamzam, M.; Zaky, I.; Elghounimy, A.; El-Naggar, S.; Magdeldin, S. Comparative Shotgun Proteomics Reveals the Characteristic Protein Signature of Osteosarcoma Subtypes. Cells 2023, 12, 2179. https://doi.org/10.3390/cells12172179
Alaa M, Al-Shehaby N, Anwar AM, Farid N, Shawky MS, Zamzam M, Zaky I, Elghounimy A, El-Naggar S, Magdeldin S. Comparative Shotgun Proteomics Reveals the Characteristic Protein Signature of Osteosarcoma Subtypes. Cells. 2023; 12(17):2179. https://doi.org/10.3390/cells12172179
Chicago/Turabian StyleAlaa, Maram, Nouran Al-Shehaby, Ali Mostafa Anwar, Nesma Farid, Mustafa Shaban Shawky, Manal Zamzam, Iman Zaky, Ahmed Elghounimy, Shahenda El-Naggar, and Sameh Magdeldin. 2023. "Comparative Shotgun Proteomics Reveals the Characteristic Protein Signature of Osteosarcoma Subtypes" Cells 12, no. 17: 2179. https://doi.org/10.3390/cells12172179
APA StyleAlaa, M., Al-Shehaby, N., Anwar, A. M., Farid, N., Shawky, M. S., Zamzam, M., Zaky, I., Elghounimy, A., El-Naggar, S., & Magdeldin, S. (2023). Comparative Shotgun Proteomics Reveals the Characteristic Protein Signature of Osteosarcoma Subtypes. Cells, 12(17), 2179. https://doi.org/10.3390/cells12172179