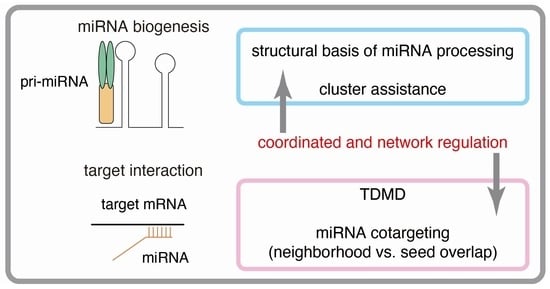
Network Regulation of microRNA Biogenesis and Target Interaction
Abstract
:1. Introduction
2. Biogenesis of Canonical miRNAs
3. Structural Basis of Pri-miRNA Processing
4. Cluster Assistance in Pri-miRNA Processing
5. Structural Basis of Pre-miRNA Processing
6. Inverse Regulation of miRNAs by Target RNAs: Target-Directed miRNA Degradation (TDMD)
7. miRNA Dosage Control by Fine-Tuning of miRNA Biogenesis Pathways
8. Impacts of Target Site Properties on Target Regulation
9. Expanded Mechanisms of Target Repression
10. Significance of miRNA Cotargeting
11. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Matsuyama, H.; Suzuki, H.I. Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis. Int. J. Mol. Sci. 2019, 21, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Luo, J.; Zhang, H.; Lu, J. microRNAs in the Same Clusters Evolve to Coordinately Regulate Functionally Related Genes. Mol. Biol. Evol. 2016, 33, 2232–2247. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.I.; Young, R.A.; Sharp, P.A. Super-Enhancer-Mediated RNA Processing Revealed by Integrative MicroRNA Network Analysis. Cell 2017, 168, 1000–1014.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.K.; Kim, B.; Kim, V.N. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E1881–E1889. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Wang, J.; Cheng, H.; Ke, X.; Sun, L.; Zhang, Q.C.; Wang, H.W. Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate. Cell 2018, 173, 1191–1203.e12. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.I.; Katsura, A.; Yasuda, T.; Ueno, T.; Mano, H.; Sugimoto, K.; Miyazono, K. Small-RNA asymmetry is directly driven by mammalian Argonautes. Nat. Struct. Mol. Biol. 2015, 22, 512–521. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Frank, F.; Sonenberg, N.; Nagar, B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 2010, 465, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Schirle, N.T.; Sheu-Gruttadauria, J.; MacRae, I.J. Structural basis for microRNA targeting. Science 2014, 346, 608–613. [Google Scholar] [CrossRef]
- Baronti, L.; Guzzetti, I.; Ebrahimi, P.; Friebe Sandoz, S.; Steiner, E.; Schlagnitweit, J.; Fromm, B.; Silva, L.; Fontana, C.; Chen, A.A.; et al. Base-pair conformational switch modulates miR-34a targeting of Sirt1 mRNA. Nature 2020, 583, 139–144. [Google Scholar] [CrossRef]
- Auyeung, V.C.; Ulitsky, I.; McGeary, S.E.; Bartel, D.P. Beyond secondary structure: Primary-sequence determinants license pri-miRNA hairpins for processing. Cell 2013, 152, 844–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, W.; Bartel, D.P. The Menu of Features that Define Primary MicroRNAs and Enable De Novo Design of MicroRNA Genes. Mol. Cell 2015, 60, 131–145. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Baek, S.C.; Lee, Y.Y.; Bastiaanssen, C.; Kim, J.; Kim, H.; Kim, V.N. A quantitative map of human primary microRNA processing sites. Mol. Cell 2021, 81, 3422–3439.e11. [Google Scholar] [PubMed]
- Kang, W.; Fromm, B.; Houben, A.J.; Hoye, E.; Bezdan, D.; Arnan, C.; Thrane, K.; Asp, M.; Johnson, R.; Biryukova, I.; et al. MapToCleave: High-throughput profiling of microRNA biogenesis in living cells. Cell Rep. 2021, 37, 110015. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Jo, M.H.; Choi, Y.G.; Park, J.; Kwon, S.C.; Hohng, S.; Kim, V.N.; Woo, J.S. Functional Anatomy of the Human Microprocessor. Cell 2015, 161, 1374–1387. [Google Scholar] [CrossRef] [Green Version]
- Herbert, K.M.; Sarkar, S.K.; Mills, M.; Delgado De la Herran, H.C.; Neuman, K.C.; Steitz, J.A. A heterotrimer model of the complete Microprocessor complex revealed by single-molecule subunit counting. RNA 2016, 22, 175–183. [Google Scholar] [CrossRef] [Green Version]
- Jin, W.; Wang, J.; Liu, C.P.; Wang, H.W.; Xu, R.M. Structural Basis for pri-miRNA Recognition by Drosha. Mol. Cell 2020, 78, 423–433.e5. [Google Scholar] [CrossRef]
- Partin, A.C.; Zhang, K.; Jeong, B.C.; Herrell, E.; Li, S.; Chiu, W.; Nam, Y. Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA. Mol. Cell 2020, 78, 411–422.e4. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.C.; Baek, S.C.; Choi, Y.G.; Yang, J.; Lee, Y.S.; Woo, J.S.; Kim, V.N. Molecular Basis for the Single-Nucleotide Precision of Primary microRNA Processing. Mol. Cell 2019, 73, 505–518.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.L.; Nguyen, T.D.; Bao, S.; Li, S.; Nguyen, T.A. The internal loops in the lower stem of primary microRNA transcripts facilitate single cleavage of human Microprocessor. Nucleic Acids Res. 2020, 48, 2579–2593. [Google Scholar] [CrossRef]
- Rice, G.M.; Shivashankar, V.; Ma, E.J.; Baryza, J.L.; Nutiu, R. Functional Atlas of Primary miRNA Maturation by the Microprocessor. Mol. Cell 2020, 80, 892–902.e4. [Google Scholar] [CrossRef] [PubMed]
- Kretov, D.A.; Walawalkar, I.A.; Mora-Martin, A.; Shafik, A.M.; Moxon, S.; Cifuentes, D. Ago2-Dependent Processing Allows miR-451 to Evade the Global MicroRNA Turnover Elicited during Erythropoiesis. Mol. Cell 2020, 78, 317–328.e6. [Google Scholar] [CrossRef] [PubMed]
- Shang, R.; Baek, S.C.; Kim, K.; Kim, B.; Kim, V.N.; Lai, E.C. Genomic Clustering Facilitates Nuclear Processing of Suboptimal Pri-miRNA Loci. Mol. Cell 2020, 78, 303–316.e4. [Google Scholar] [CrossRef]
- Fang, W.; Bartel, D.P. MicroRNA Clustering Assists Processing of Suboptimal MicroRNA Hairpins through the Action of the ERH Protein. Mol. Cell 2020, 78, 289–302.e6. [Google Scholar] [CrossRef]
- Kwon, S.C.; Jang, H.; Shen, S.; Baek, S.C.; Kim, K.; Yang, J.; Kim, J.; Kim, J.S.; Wang, S.; Shi, Y.; et al. ERH facilitates microRNA maturation through the interaction with the N-terminus of DGCR8. Nucleic Acids Res. 2020, 48, 11097–11112. [Google Scholar] [CrossRef]
- Hutter, K.; Lohmuller, M.; Jukic, A.; Eichin, F.; Avci, S.; Labi, V.; Szabo, T.G.; Hoser, S.M.; Huttenhofer, A.; Villunger, A.; et al. SAFB2 Enables the Processing of Suboptimal Stem-Loop Structures in Clustered Primary miRNA Transcripts. Mol. Cell 2020, 78, 876–889.e6. [Google Scholar]
- Vilimova, M.; Contrant, M.; Randrianjafy, R.; Dumas, P.; Elbasani, E.; Ojala, P.M.; Pfeffer, S.; Fender, A. Cis regulation within a cluster of viral microRNAs. Nucleic Acids Res. 2021, 49, 10018–10033. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Plessmann, U.; Harlander, S.; Daiss, J.L.; Eichner, N.; Lehmann, G.; Schall, K.; Urlaub, H.; Meister, G. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol. Cell 2017, 66, 270–284.e13. [Google Scholar] [CrossRef] [PubMed]
- Nussbacher, J.K.; Yeo, G.W. Systematic Discovery of RNA Binding Proteins that Regulate MicroRNA Levels. Mol. Cell 2018, 69, 1005–1016.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, P.; Wang, L.; Sliz, P.; Gregory, R.I. A Biogenesis Step Upstream of Microprocessor Controls miR-17 approximately 92 Expression. Cell 2015, 162, 885–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donayo, A.O.; Johnson, R.M.; Tseng, H.W.; Izreig, S.; Gariepy, A.; Mayya, V.K.; Wu, E.; Alam, R.; Lussier, C.; Jones, R.G.; et al. Oncogenic Biogenesis of pri-miR-17 approximately 92 Reveals Hierarchy and Competition among Polycistronic MicroRNAs. Mol. Cell 2019, 75, 340–356.e10. [Google Scholar] [CrossRef] [PubMed]
- Zapletal, D.; Taborska, E.; Pasulka, J.; Malik, R.; Kubicek, K.; Zanova, M.; Much, C.; Sebesta, M.; Buccheri, V.; Horvat, F.; et al. Structural and functional basis of mammalian microRNA biogenesis by Dicer. Mol. Cell 2022, 82, 4064–4079.e13. [Google Scholar] [CrossRef]
- Jouravleva, K.; Golovenko, D.; Demo, G.; Dutcher, R.C.; Hall, T.M.T.; Zamore, P.D.; Korostelev, A.A. Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB. Mol. Cell 2022, 82, 4049–4063.e6. [Google Scholar] [CrossRef]
- Su, S.; Wang, J.; Deng, T.; Yuan, X.; He, J.; Liu, N.; Li, X.; Huang, Y.; Wang, H.W.; Ma, J. Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD. Nature 2022, 607, 399–406. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Naganuma, M.; Nishizawa, T.; Kusakizako, T.; Tomari, Y.; Nishimasu, H.; Nureki, O. Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex. Nature 2022, 607, 393–398. [Google Scholar] [CrossRef]
- Park, J.E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef] [Green Version]
- Heo, I.; Ha, M.; Lim, J.; Yoon, M.J.; Park, J.E.; Kwon, S.C.; Chang, H.; Kim, V.N. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 2012, 151, 521–532. [Google Scholar] [CrossRef] [Green Version]
- Gu, S.; Jin, L.; Zhang, Y.; Huang, Y.; Zhang, F.; Valdmanis, P.N.; Kay, M.A. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell 2012, 151, 900–911. [Google Scholar] [CrossRef]
- Tsutsumi, A.; Kawamata, T.; Izumi, N.; Seitz, H.; Tomari, Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat. Struct. Mol. Biol. 2011, 18, 1153–1158. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, J.; Li, G.; Wang, H.W. Structure of precursor microRNA’s terminal loop regulates human Dicer’s dicing activity by switching DExH/D domain. Protein Cell 2015, 6, 185–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zeng, Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010, 38, 7689–7697. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.D.; Trinh, T.A.; Bao, S.; Nguyen, T.A. Secondary structure RNA elements control the cleavage activity of DICER. Nat. Commun. 2022, 13, 2138. [Google Scholar]
- Reichholf, B.; Herzog, V.A.; Fasching, N.; Manzenreither, R.A.; Sowemimo, I.; Ameres, S.L. Time-Resolved Small RNA Sequencing Unravels the Molecular Principles of MicroRNA Homeostasis. Mol. Cell 2019, 75, 756–768.e7. [Google Scholar] [CrossRef]
- Kingston, E.R.; Bartel, D.P. Global analyses of the dynamics of mammalian microRNA metabolism. Genome Res. 2019, 29, 1777–1790. [Google Scholar] [CrossRef]
- De, N.; Young, L.; Lau, P.W.; Meisner, N.C.; Morrissey, D.V.; MacRae, I.J. Highly complementary target RNAs promote release of guide RNAs from human Argonaute2. Mol. Cell 2013, 50, 344–355. [Google Scholar] [CrossRef] [Green Version]
- Jo, M.H.; Shin, S.; Jung, S.R.; Kim, E.; Song, J.J.; Hohng, S. Human Argonaute 2 Has Diverse Reaction Pathways on Target RNAs. Mol. Cell 2015, 59, 117–124. [Google Scholar] [CrossRef] [Green Version]
- Park, J.H.; Shin, S.Y.; Shin, C. Non-canonical targets destabilize microRNAs in human Argonautes. Nucleic Acids Res. 2017, 45, 1569–1583. [Google Scholar] [CrossRef]
- Ameres, S.L.; Horwich, M.D.; Hung, J.H.; Xu, J.; Ghildiyal, M.; Weng, Z.; Zamore, P.D. Target RNA-directed trimming and tailing of small silencing RNAs. Science 2010, 328, 1534–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cazalla, D.; Yario, T.; Steitz, J.A. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 2010, 328, 1563–1566. [Google Scholar] [CrossRef] [PubMed]
- Piwecka, M.; Glazar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleaveland, B.; Shi, C.Y.; Stefano, J.; Bartel, D.P. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 2018, 174, 350–362.e17. [Google Scholar] [CrossRef] [Green Version]
- Bitetti, A.; Mallory, A.C.; Golini, E.; Carrieri, C.; Carreno Gutierrez, H.; Perlas, E.; Perez-Rico, Y.A.; Tocchini-Valentini, G.P.; Enright, A.J.; Norton, W.H.J.; et al. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat. Struct. Mol. Biol. 2018, 25, 244–251. [Google Scholar] [CrossRef]
- Donnelly, B.F.; Yang, B.; Grimme, A.L.; Vieux, K.F.; Liu, C.Y.; Zhou, L.; McJunkin, K. The developmentally timed decay of an essential microRNA family is seed-sequence dependent. Cell Rep. 2022, 40, 111154. [Google Scholar] [CrossRef]
- Kingston, E.R.; Blodgett, L.W.; Bartel, D.P. Endogenous transcripts direct microRNA degradation in Drosophila, and this targeted degradation is required for proper embryonic development. Mol. Cell 2022, 82, 3872–3884.e9. [Google Scholar] [CrossRef]
- Duan, Y.; Veksler-Lublinsky, I.; Ambros, V. Critical contribution of 3′ non-seed base pairing to the in vivo function of the evolutionarily conserved let-7a microRNA. Cell Rep. 2022, 39, 110745. [Google Scholar] [CrossRef]
- Shi, C.Y.; Kingston, E.R.; Kleaveland, B.; Lin, D.H.; Stubna, M.W.; Bartel, D.P. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science 2020, 370, eabc9359. [Google Scholar] [CrossRef]
- Han, J.; LaVigne, C.A.; Jones, B.T.; Zhang, H.; Gillett, F.; Mendell, J.T. A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science 2020, 370, eabc9546. [Google Scholar] [CrossRef]
- Sheu-Gruttadauria, J.; Pawlica, P.; Klum, S.M.; Wang, S.; Yario, T.A.; Schirle Oakdale, N.T.; Steitz, J.A.; MacRae, I.J. Structural Basis for Target-Directed MicroRNA Degradation. Mol. Cell 2019, 75, 1243–1255.e7. [Google Scholar] [CrossRef]
- Li, L.; Sheng, P.; Li, T.; Fields, C.J.; Hiers, N.M.; Wang, Y.; Li, J.; Guardia, C.M.; Licht, J.D.; Xie, M. Widespread microRNA degradation elements in target mRNAs can assist the encoded proteins. Genes Dev. 2021, 35, 1595–1609. [Google Scholar] [CrossRef] [PubMed]
- Ghini, F.; Rubolino, C.; Climent, M.; Simeone, I.; Marzi, M.J.; Nicassio, F. Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat. Commun. 2018, 9, 3119. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Mendell, J.T. MicroRNA turnover: A tale of tailing, trimming, and targets. Trends Biochem. Sci. 2022, 48, 26–39. [Google Scholar] [CrossRef]
- Martello, G.; Rosato, A.; Ferrari, F.; Manfrin, A.; Cordenonsi, M.; Dupont, S.; Enzo, E.; Guzzardo, V.; Rondina, M.; Spruce, T.; et al. A MicroRNA targeting dicer for metastasis control. Cell 2010, 141, 1195–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Pedersen, J.S.; Kwon, S.C.; Belair, C.D.; Kim, Y.K.; Yeom, K.H.; Yang, W.Y.; Haussler, D.; Blelloch, R.; Kim, V.N. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 2009, 136, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Lyu, X.; Ding, L.; Ke, L.; Yang, D.; Pirouz, M.; Qi, Y.; Ong, J.; Gao, G.; Du, P.; et al. Global miRNA dosage control of embryonic germ layer specification. Nature 2021, 593, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Li, X.; Pritykin, Y.; Concepcion, C.P.; Lu, Y.; La Rocca, G.; Zhang, M.; King, B.; Cook, P.J.; Au, Y.W.; Popow, O.; et al. High-Resolution In Vivo Identification of miRNA Targets by Halo-Enhanced Ago2 Pull-Down. Mol. Cell 2020, 79, 167–179.e11. [Google Scholar] [CrossRef]
- Patel, R.K.; West, J.D.; Jiang, Y.; Fogarty, E.A.; Grimson, A. Robust partitioning of microRNA targets from downstream regulatory changes. Nucleic Acids Res. 2020, 48, 9724–9746. [Google Scholar] [CrossRef]
- Kern, F.; Krammes, L.; Danz, K.; Diener, C.; Kehl, T.; Kuchler, O.; Fehlmann, T.; Kahraman, M.; Rheinheimer, S.; Aparicio-Puerta, E.; et al. Validation of human microRNA target pathways enables evaluation of target prediction tools. Nucleic Acids Res. 2021, 49, 127–144. [Google Scholar] [CrossRef] [PubMed]
- Serra, F.; Bottini, S.; Pratella, D.; Stathopoulou, M.G.; Sebille, W.; El-Hami, L.; Repetto, E.; Mauduit, C.; Benahmed, M.; Grandjean, V.; et al. Systemic CLIP-seq analysis and game theory approach to model microRNA mode of binding. Nucleic Acids Res. 2021, 49, e66. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, X.; Cai, Z.; Zhou, J.; Cao, R.; Zhao, Y.; Chen, Z.; Wang, D.; Ruan, W.; Zhao, Q.; et al. A novel class of microRNA-recognition elements that function only within open reading frames. Nat. Struct. Mol. Biol. 2018, 25, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Bofill-De Ros, X.; Shao, T.J.; Jiang, M.; Li, K.; Villanueva, P.; Dai, L.; Gu, S. 3′ Uridylation Confers miRNAs with Non-canonical Target Repertoires. Mol. Cell 2019, 75, 511–522.e4. [Google Scholar] [CrossRef]
- Kelly, T.J.; Brummer, A.; Hooshdaran, N.; Tariveranmoshabad, M.; Zamudio, J.R. Temporal Control of the TGF-beta Signaling Network by Mouse ESC MicroRNA Targets of Different Affinities. Cell Rep. 2019, 29, 2702–2717.e7. [Google Scholar] [CrossRef]
- Schmiedel, J.M.; Klemm, S.L.; Zheng, Y.; Sahay, A.; Bluthgen, N.; Marks, D.S.; van Oudenaarden, A. Gene expression. MicroRNA control of protein expression noise. Science 2015, 348, 128–132. [Google Scholar] [CrossRef]
- Wei, L.; Li, S.; Zhang, P.; Hu, T.; Zhang, M.Q.; Xie, Z.; Wang, X. Characterizing microRNA-mediated modulation of gene expression noise and its effect on synthetic gene circuits. Cell Rep. 2021, 36, 109573. [Google Scholar] [CrossRef] [PubMed]
- Svoronos, A.A.; Campbell, S.G.; Engelman, D.M. MicroRNA function can be reversed by altering target gene expression levels. iScience 2021, 24, 103208. [Google Scholar] [CrossRef]
- Becker, W.R.; Ober-Reynolds, B.; Jouravleva, K.; Jolly, S.M.; Zamore, P.D.; Greenleaf, W.J. High-Throughput Analysis Reveals Rules for Target RNA Binding and Cleavage by AGO2. Mol. Cell 2019, 75, 741–755.e11. [Google Scholar] [CrossRef]
- Eisen, T.J.; Eichhorn, S.W.; Subtelny, A.O.; Bartel, D.P. MicroRNAs Cause Accelerated Decay of Short-Tailed Target mRNAs. Mol. Cell 2020, 77, 775–785.e8. [Google Scholar] [CrossRef]
- Baek, D.; Villen, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [Green Version]
- Bazzini, A.A.; Lee, M.T.; Giraldez, A.J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012, 336, 233–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biasini, A.; Abdulkarim, B.; de Pretis, S.; Tan, J.Y.; Arora, R.; Wischnewski, H.; Dreos, R.; Pelizzola, M.; Ciaudo, C.; Marques, A.C. Translation is required for miRNA-dependent decay of endogenous transcripts. EMBO J. 2021, 40, e104569. [Google Scholar] [CrossRef]
- Ruijtenberg, S.; Sonneveld, S.; Cui, T.J.; Logister, I.; de Steenwinkel, D.; Xiao, Y.; MacRae, I.J.; Joo, C.; Tanenbaum, M.E. mRNA structural dynamics shape Argonaute-target interactions. Nat. Struct. Mol. Biol. 2020, 27, 790–801. [Google Scholar] [CrossRef]
- Kim, S.; Kim, S.; Chang, H.R.; Kim, D.; Park, J.; Son, N.; Park, J.; Yoon, M.; Chae, G.; Kim, Y.K.; et al. The regulatory impact of RNA-binding proteins on microRNA targeting. Nat. Commun. 2021, 12, 5057. [Google Scholar] [CrossRef] [PubMed]
- Grigelioniene, G.; Suzuki, H.I.; Taylan, F.; Mirzamohammadi, F.; Borochowitz, Z.U.; Ayturk, U.M.; Tzur, S.; Horemuzova, E.; Lindstrand, A.; Weis, M.A.; et al. Gain-of-function mutation of microRNA-140 in human skeletal dysplasia. Nat. Med. 2019, 25, 583–590. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.I.; Spengler, R.M.; Grigelioniene, G.; Kobayashi, T.; Sharp, P.A. Deconvolution of seed and RNA-binding protein crosstalk in RNAi-based functional genomics. Nat. Genet. 2018, 50, 657–661. [Google Scholar] [CrossRef]
- Kelly, T.J.; Suzuki, H.I.; Zamudio, J.R.; Suzuki, M.; Sharp, P.A. Sequestration of microRNA-mediated target repression by the Ago2-associated RNA-binding protein FAM120A. RNA 2019, 25, 1291–1297. [Google Scholar] [CrossRef]
- Doench, J.G.; Sharp, P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004, 18, 504–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Hon, L.S.; Zhang, Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol. 2007, 8, R166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saetrom, P.; Heale, B.S.; Snove, O., Jr.; Aagaard, L.; Alluin, J.; Rossi, J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007, 35, 2333–2342. [Google Scholar] [CrossRef]
- Briskin, D.; Wang, P.Y.; Bartel, D.P. The biochemical basis for the cooperative action of microRNAs. Proc. Natl. Acad. Sci. USA 2020, 117, 17764–17774. [Google Scholar] [CrossRef]
- Whipple, A.J.; Breton-Provencher, V.; Jacobs, H.N.; Chitta, U.K.; Sur, M.; Sharp, P.A. Imprinted Maternally Expressed microRNAs Antagonize Paternally Driven Gene Programs in Neurons. Mol. Cell 2020, 78, 85–95.e8. [Google Scholar] [CrossRef]
- Cantini, L.; Bertoli, G.; Cava, C.; Dubois, T.; Zinovyev, A.; Caselle, M.; Castiglioni, I.; Barillot, E.; Martignetti, L. Identification of microRNA clusters cooperatively acting on epithelial to mesenchymal transition in triple negative breast cancer. Nucleic Acids Res. 2019, 47, 2205–2215. [Google Scholar] [CrossRef] [Green Version]
- Jin, W.; Mulas, F.; Gaertner, B.; Sui, Y.; Wang, J.; Matta, I.; Zeng, C.; Vinckier, N.; Wang, A.; Nguyen-Ngoc, K.V.; et al. A Network of microRNAs Acts to Promote Cell Cycle Exit and Differentiation of Human Pancreatic Endocrine Cells. iScience 2019, 21, 681–694. [Google Scholar] [CrossRef] [Green Version]
- Cherone, J.M.; Jorgji, V.; Burge, C.B. Cotargeting among microRNAs in the brain. Genome Res. 2019, 29, 1791–1804. [Google Scholar] [CrossRef]
- Kitai, H.; Kato, N.; Ogami, K.; Komatsu, S.; Watanabe, Y.; Yoshino, S.; Koshi, E.; Tsubota, S.; Funahashi, Y.; Maeda, T.; et al. Systematic characterization of seed overlap microRNA cotargeting associated with lupus pathogenesis. BMC Biol. 2022, 20, 248. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komatsu, S.; Kitai, H.; Suzuki, H.I. Network Regulation of microRNA Biogenesis and Target Interaction. Cells 2023, 12, 306. https://doi.org/10.3390/cells12020306
Komatsu S, Kitai H, Suzuki HI. Network Regulation of microRNA Biogenesis and Target Interaction. Cells. 2023; 12(2):306. https://doi.org/10.3390/cells12020306
Chicago/Turabian StyleKomatsu, Shintaro, Hiroki Kitai, and Hiroshi I. Suzuki. 2023. "Network Regulation of microRNA Biogenesis and Target Interaction" Cells 12, no. 2: 306. https://doi.org/10.3390/cells12020306
APA StyleKomatsu, S., Kitai, H., & Suzuki, H. I. (2023). Network Regulation of microRNA Biogenesis and Target Interaction. Cells, 12(2), 306. https://doi.org/10.3390/cells12020306