Social Isolation Activates Dormant Mammary Tumors, and Modifies Inflammatory and Mitochondrial Metabolic Pathways in the Rat Mammary Gland
Abstract
:1. Introduction
2. Methods
2.1. Animals
2.2. Mammary Tumor Induction and Social Isolation
2.3. Tamoxifen Therapy and Administration of Jaeumganghwa-Tang (JGT)
2.4. Monitoring Mammary Tumor Responses
2.5. Mammary Gland and Tumor Harvesting
2.6. Tumor Pathologic Evaluation
2.7. RNA-Sequencing
2.8. RNA-Seq Data Analysis
2.9. Knowledge-Guided Differential Dependency Network (KDDN) Analysis
2.10. Quantitative Real-Time Polymerase Chain Reaction
2.11. Protein Extraction and Western Blotting
2.12. Lactate Dehydrogenase (LDH) Activity and Lactate Level Assays
2.13. Statistical Analysis
3. Results
3.1. Social Isolation Does Not Modify Tamoxifen Responsiveness during Therapy
3.2. JGT Increases Responsiveness to Tamoxifen
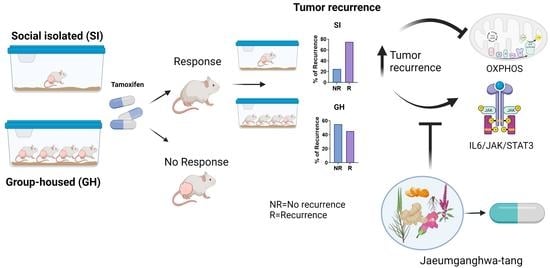
3.3. Social Isolation Increases the Risk of the Regrowth of Dormant Mammary Tumors and Local Recurrence in Rats, and JGT Prevents this Increase
3.4. Tamoxifen and JGT Modify Tumor Histopathology
3.5. Social Isolation and JGT Modify IL6/JAK/STAT3 and Oxidative Phosphorylation Signaling in Mammary Glands and Tumors
3.5.1. Effects of Social Isolation
3.5.2. Effects of JGT on Socially Isolated Rats after Tamoxifen Treatment
3.5.3. Genes Altered by Social Isolation and Reversed JGT in the IL6/JAK/STAT3 and OXPHOS Pathways
IL6/JAK/STAT3 Pathway
OXPHOS Pathway
3.5.4. Effects of JGT on the Tamoxifen-Treated Group-Housed Rats
3.5.5. Knowledge-Fused Differential Dependency Network (KDDN) Analysis
3.6. Verification of Differential Gene Expression
3.6.1. Inflammatory Genes
3.6.2. OXPHOS Genes
3.6.3. Effect of JGT on Tamoxifen-Treated GH Rats
3.6.4. IL6 and STAT3 Expression in Mammary Tumors
3.7. Social Isolation Upregulates Receptors for RAGE in Mammary Glands and Tumors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rehkopf, D.; Jutte, D.; Syme, S.L.; Balmes, J.; Adler, N. Social Isolation: A Predictor of Mortality Comparable to Traditional Clinical Risk Factors. Am. J. Public Health 2013, 103, 2056–2062. [Google Scholar]
- Hakulinen, C.; Pulkki-Råback, L.; Virtanen, M.; Jokela, M.; Kivimaki, M.; Elovainio, M. Social isolation and loneliness as risk factors for myocardial infarction, stroke and mortality: UK Biobank cohort study of 479 054 men and women. Heart 2018, 104, 1536–1542. [Google Scholar] [CrossRef]
- Brinkhues, S.; Dukers-Muijrers, N.H.T.M.; Hoebe, C.J.P.A.; Van Der Kallen, C.J.H.; Dagnelie, P.C.; Koster, A.; Henry, R.M.A.; Sep, S.J.S.; Schaper, N.C.; Stehouwer, C.D.A.; et al. Socially isolated individuals are more prone to have newly diagnosed and prevalent type 2 diabetes mellitus—the Maastricht study. BMC Public Health 2017, 17, 955. [Google Scholar] [CrossRef] [Green Version]
- Penninkilampi, R.; Casey, A.-N.; Singh, M.F.; Brodaty, H. The Association between Social Engagement, Loneliness, and Risk of Dementia: A Systematic Review and Meta-Analysis. J. Alzheimer’s Dis. 2018, 66, 1619–1633. [Google Scholar] [CrossRef]
- Subramanian, I.; Farahnik, J.; Mischley, L.K. Synergy of pandemics-social isolation is associated with worsened Parkinson severity and quality of life. npj Park. Dis. 2020, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Alcaraz, K.; Eddens, K.S.; Blase, J.L.; Diver, W.R.; Patel, A.V.; Teras, L.R.; Stevens, V.L.; Jacobs, E.J.; Gapstur, S.M. Social Isolation and Mortality in US Black and White Men and Women. Am. J. Epidemiol. 2018, 188, 102–109. [Google Scholar] [CrossRef] [Green Version]
- Pinquart, M.; Duberstein, P.R. Associations of social networks with cancer mortality: A meta-analysis. Crit. Rev. Oncol. 2010, 75, 122–137. [Google Scholar] [CrossRef] [Green Version]
- Murthy, V.H.; Murthy, V.H. Together; Harper Collins Publishers: New York, NY, USA, 2020. [Google Scholar]
- Smith, B.; Lim, M. How the COVID-19 pandemic is focusing attention on loneliness and social isolation. Public Health Res. Pr. 2020, 30, 3022008. [Google Scholar] [CrossRef] [PubMed]
- Kroenke, C.H.; Michael, Y.L.; Poole, E.M.; Kwan, M.L.; Nechuta, S.; Leas, E.; Caan, B.J.; Pierce, J.; Shu, X.-O.; Zheng, Y.; et al. Postdiagnosis social networks and breast cancer mortality in the After Breast Cancer Pooling Project. Cancer 2016, 123, 1228–1237. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, A.; Kawachi, I.; Iso, H.; Iwasaki, M.; Inoue, M.; Tsugane, S. Social support and cancer incidence and mortality: The JPHC study cohort II. Cancer Causes Control. 2013, 24, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Kroenke, C.H.; Kubzansky, L.D.; Schernhammer, E.S.; Holmes, M.D.; Kawachi, I. Social networks, social support, and sur-vival after breast cancer diagnosis. J. Clin. Oncol. 2006, 24, 1105–1111. Available online: http://www.ncbi.nlm.nih.gov/pubmed/16505430 (accessed on 10 April 2022). [CrossRef] [PubMed]
- Kroemer, G.; McQuade, J.L.; Merad, M.; André, F.; Zitvogel, L. Bodywide ecological interventions on cancer. Nat. Med. 2023, 29, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Leschak, C.J.M.; Eisenberger, N.I. Two Distinct Immune Pathways Linking Social Relationships With Health: Inflammatory and Antiviral Processes. Psychosom. Med. 2019, 81, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.J.; Gavey, S.; Riddell, N.E.; Kontari, P.; Victor, C. The association between loneliness, social isolation and inflammation: A systematic review and meta-analysis. Neurosci. Biobehav. Rev. 2020, 112, 519–541. [Google Scholar] [CrossRef]
- Sun, M.; Choi, E.Y.; Magee, D.J.; Stets, C.W.; During, M.J.; Lin, E.-J.D. Metabolic Effects of Social Isolation in Adult C57BL/6 Mice. Int. Sch. Res. Not. 2014, 2014, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zhuravliova, E.; Barbakadze, T.; Zaalishvili, E.; Chipashvili, M.; Koshoridze, N.; Mikeladze, D. Social isolation in rats inhibits oxidative metabolism, decreases the content of mitochondrial K-Ras and activates mitochondrial hexokinase. Behav. Brain Res. 2009, 205, 377–383. [Google Scholar] [CrossRef]
- Sonei, N.; Amiri, S.; Jafarian, I.; Anoush, M.; Rahimi-Balaei, M.; Bergen, H.; Haj-Mirzaian, A.; Hosseini, M.-J. Mitochondrial dysfunction bridges negative affective disorders and cardiomyopathy in socially isolated rats: Pros and cons of fluoxetine. World J. Biol. Psychiatry 2016, 18, 39–53. [Google Scholar] [CrossRef]
- Andrade, F.D.O.; Yu, W.; Zhang, X.; Carney, E.; Hu, R.; Clarke, R.; FitzGerald, K.; Hilakivi-Clarke, L. Effects of Jaeumkanghwa-tang on tamoxifen responsiveness in preclinical ER+ breast cancer model. Endocrine-Related Cancer 2019, 26, 339–353. [Google Scholar] [CrossRef]
- Kim, Y.K.; Kim, H.J.; Kim, W.S.; Park, H.J.; Moon, G.; Kim, D.W.; Won, J.H. Inhibitory effect of jaeumgamghwa-tang on al-lergic inflammatory reaction. J. Korean Orient. Intern. Med. 2004, 25, 174–182. [Google Scholar] [CrossRef]
- Lee, S.H.; Yang, E.J. Anti-Neuroinflammatory Effect of Jaeumganghwa-Tang in an Animal Model of Amyotrophic Lateral Sclerosis. Evidence-Based Complement. Altern. Med. 2019, 2019, 1–7. [Google Scholar] [CrossRef]
- Kim, A.; Im, M.; Hwang, Y.-H.; Yang, H.J.; Ma, J.Y. Jaeumganghwa-Tang Induces Apoptosis via the Mitochondrial Pathway and Lactobacillus Fermentation Enhances Its Anti-Cancer Activity in HT1080 Human Fibrosarcoma Cells. PLoS ONE 2015, 10, e0127898. [Google Scholar] [CrossRef] [Green Version]
- Che, C.-T.; Wang, Z.J.; Chow, M.S.S.; Lam, C.W.K. Herb-Herb Combination for Therapeutic Enhancement and Advancement: Theory, Practice and Future Perspectives. Molecules 2013, 18, 5125–5141. [Google Scholar] [CrossRef] [Green Version]
- Hermes, G.L.; Delgado, B.; Tretiakova, M.; Cavigelli, S.A.; Krausz, T.; Conzen, S.D.; McClintock, M.K. Social isolation dysregulates endocrine and behavioral stress while increasing malignant burden of spontaneous mammary tumors. Proc. Natl. Acad. Sci. USA 2009, 106, 22393–22398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, J.B.; Pang, D.; Delgado, B.; Kocherginsky, M.; Tretiakova, M.; Krausz, T.; Pan, D.; He, J.; McClintock, M.K.; Conzen, S.D. A Model of Gene-Environment Interaction Reveals Altered Mammary Gland Gene Expression and Increased Tumor Growth following Social Isolation. Cancer Prev. Res. 2009, 2, 850–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, A.; Collacchi, B.; Capoccia, S.; D’Urso, M.T.; Cecchetti, S.; Raggi, C.; Sestili, P.; Aricò, E.; Pontecorvi, G.; Puglisi, R.; et al. Chronic Isolation Stress Affects Central Neuroendocrine Signaling Leading to a Metabolically Active Microenvironment in a Mouse Model of Breast Cancer. Front. Behav. Neurosci. 2021, 15, 660738. [Google Scholar] [CrossRef]
- Marchant, J. The effects of different social conditions on breast cancer induction in three genetic types of mice by dibenz[a,h]anthracene and a comparison with breast carcinogenesis by 3-methylcholanthrene. Br. J. Cancer 1967, 21, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Sumis, A.; Cook, K.L.; Andrade, A.F.F.; Hu, R.; Kidney, E.; Zhang, X.; Kim, D.; Carney, E.; Nguyen, N.; Yu, W.; et al. Social isolation induces unfolded protein response and autophagy in the mouse mammary gland: Link to obesity and mammary cancer risk. Endocr. Relat. Cancer 2016, 10, 839–856. [Google Scholar]
- Qin, J.-F.; Jin, F.-J.; Li, N.; Guan, H.-T.; Lan, L.; Ni, H.; Wang, Y. Adrenergic receptor β2 activation by stress promotes breast cancer progression through macrophages M2 polarization in tumor microenvironment. BMB Rep. 2015, 48, 295–300. [Google Scholar] [CrossRef] [Green Version]
- Budiu, R.A.; Vlad, A.M.; Nazario, L.; Bathula, C.; Cooper, K.L.; Edmed, J.; Thaker, P.H.; Urban, J.; Kalinski, P.; Lee, A.V.; et al. Restraint and Social Isolation Stressors Differentially Regulate Adaptive Immunity and Tumor Angiogenesis in a Breast Cancer Mouse Model. Cancer Clin. Oncol. 2016, 6, p12. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korsh, J.; Shen, A.; Aliano, K.; Davenport, T. Polycyclic Aromatic Hydrocarbons and Breast Cancer: A Review of the Literature. Breast Care 2015, 10, 316–318. [Google Scholar] [CrossRef] [Green Version]
- Madden, K.S.; Szpunar, M.J.; Brown, E.B. Early impact of social isolation and breast tumor progression in mice. Brain Behav. Immun. 2012, 30, S135–S141. [Google Scholar] [CrossRef] [Green Version]
- Hilakivi-Clarke, L.; Wärri, A.; Bouker, K.B.; Zhang, X.; Cook, K.L.; Jin, L.; Zwart, A.; Nguyen, N.; Hu, R.; Cruz, M.I.; et al. Effects of In Utero Exposure to Ethinyl Estradiol on Tamoxifen Resistance and Breast Cancer Recurrence in a Preclinical Model. Gynecol. Oncol. 2016, 109. [Google Scholar] [CrossRef] [PubMed]
- Reagan-Shaw, S.; Nihal, M.; Ahmad, N. Dose translation from animal to human studies revisited. FASEB J. 2008, 22, 659–661. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.M.; Lee, Y.W.; Yoo, H.S.; Cho, C.K. Case study of a breast cancer patient accompanying with hot flush by tamoxi-fen whose condition was improved by Jayeumganghwa-tang. Korean J. Orient. Int. Med. 2010, 31, 395–400. [Google Scholar]
- Zhang, X.; Cook, K.L.; Warri, A.; Cruz, I.M.; Rosim, M.; Riskin, J.; Helferich, W.; Doerge, D.; Clarke, R.; Hilakivi-Clarke, L. Lifetime Genistein Intake Increases the Response of Mammary Tumors to Tamoxifen in Rats. Clin. Cancer Res. 2017, 23, 814–824. [Google Scholar] [CrossRef] [Green Version]
- Fisher, B.; Costantino, J.P.; Wickerham, D.L.; Redmond, C.K.; Kavanah, M.; Cronin, W.M.; Vogel, V.; Robidoux, A.; Dimitrov, N.; Atkins, J.; et al. Tamoxifen for Prevention of Breast Cancer: Report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. Gynecol. Oncol. 1998, 90, 1371–1388. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Mofers, A.; Hydbring, P.; Olofsson, M.H.; Guo, J.; Linder, S.; D’Arcy, P. MYC is downregulated by a mitochondrial checkpoint mechanism. Oncotarget 2017, 8, 90225–90237. [Google Scholar] [CrossRef] [Green Version]
- Martin, T.R.; Wurfel, M.M.; Zanoni, I.; Ulevitch, R. Targeting innate immunity by blocking CD14: Novel approach to control inflammation and organ dysfunction in COVID-19 illness. Ebiomedicine 2020, 57, 102836. [Google Scholar] [CrossRef]
- Zhang, N.; Zhao, Y.D.; Wang, X.M. CXCL10 an important chemokine associated with cytokine storm in COVID-19 infected patients. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7497–7505. [Google Scholar] [PubMed]
- Zhao, Y.; Kilian, C.; Turner, J.-E.; Bosurgi, L.; Roedl, K.; Bartsch, P.; Gnirck, A.-C.; Cortesi, F.; Schultheiß, C.; Hellmig, M.; et al. Clonal expansion and activation of tissue-resident memory-like T H 17 cells expressing GM-CSF in the lungs of patients with severe COVID-19. Sci. Immunol. 2021, 6. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, B.; Hoffman, E.P.; Clarke, R.; Zhang, Z.; Shih, I.-M.; Xuan, J.; Herrington, D.M.; Wang, Y. KDDN: An open-source Cytoscape app for constructing differential dependency networks with significant rewiring. Bioinformatics 2014, 31, 287–289. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.; Zhao, Y.; Becker, M.; John, S.; Parekh, B.S.; Huang, S.; Hendarwanto, A.; Martinez, E.D.; Chen, Y.; Lu, H.; et al. HDAC1 Acetylation Is Linked to Progressive Modulation of Steroid Receptor-Induced Gene Transcription. Mol. Cell 2006, 22, 669–679. [Google Scholar] [CrossRef]
- Ghazawi, F.M.; Faller, E.M.; Parmar, P.; El-Salfiti, A.; MacPherson, P.A. Suppressor of cytokine signaling (SOCS) proteins are induced by IL-7 and target surface CD127 protein for degradation in human CD8 T cells. Cell. Immunol. 2016, 306-307, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Svensson, M.N.; Doody, K.M.; Schmiedel, B.J.; Bhattacharyya, S.; Panwar, B.; Wiede, F.; Yang, S.; Santelli, E.; Wu, D.J.; Sacchetti, C.; et al. Reduced expression of phosphatase PTPN2 promotes pathogenic conversion of Tregs in autoimmunity. J. Clin. Investig. 2019, 129, 1193–1210. [Google Scholar] [CrossRef] [Green Version]
- Koc, E.C.; Kartal, F.; Tirona, M.; Koc, H. Reduced Mitochondrial Ribosomal Protein Expression impairs Oxidative Phosphorylation and Apoptosis in ER/PR(+) Breast Cancer Cell lines. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Coll, R.C.; Holley, C.L.; Schroder, K. Mitochondrial DNA synthesis fuels NLRP3 inflammasome. Cell Res. 2018, 28, 1046–1047. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Méndez, J.D.; Méndez-Valenzuela, V.; Hernandez, M.M.A. Cellular signalling of the receptor for advanced glycation end products (RAGE). Cell. Signal. 2013, 25, 2185–2197. [Google Scholar] [CrossRef]
- Uchino, B.N.; Trettevik, R.; de Grey, R.G.K.; Cronan, S.; Hogan, J.; Baucom, B.R.W. Social support, social integration, and inflammatory cytokines: A meta-analysis. Health Psychol. 2018, 37, 462–471. [Google Scholar] [CrossRef]
- Banerjee, K.; Resat, H. Constitutive activation of STAT3 in breast cancer cells: A review. Int. J. Cancer 2015, 138, 2570–2578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Bankaitis, K.V.; Fingleton, B. Targeting IL4/IL4R for the treatment of epithelial cancer metastasis. Clin. Exp. Metastasis 2015, 32, 847–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, X.; Kim, J.; Deng, M.; John, S.; Chen, H.; Wu, G.; Phan, H.; Zhang, C.C. Inhibitory leukocyte immunoglobulin-like receptors: Immune checkpoint proteins and tumor sustaining factors. Cell Cycle 2016, 15, 25–40. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, J.; Wu, W.; Gao, H.; Liu, N.; Zhan, G.; Li, L.; Han, L.; Guo, X. Myeloid-derived suppressor cells promote epithelial ovarian cancer cell stemness by inducing the CSF2/p-STAT3 signalling pathway. FEBS J. 2020, 287, 5218–5235. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Cheon, S.; Cho, D. The dual effects of interleukin-18 in tumor progression. Cell. Mol. Immunol. 2007, 4, 329–335. [Google Scholar]
- Avagliano, A.; Ruocco, M.R.; Aliotta, F.; Belviso, I.; Accurso, A.; Masone, S.; Montagnani, S.; Arcucci, A. Mitochondrial Flexibility of Breast Cancers: A Growth Advantage and a Therapeutic Opportunity. Cells 2019, 8, 401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barlow, J.; Solomon, T.P.J.; Affourtit, C. Pro-inflammatory cytokines attenuate glucose-stimulated insulin secretion from INS-1E insulinoma cells by restricting mitochondrial pyruvate oxidation capacity—Novel mechanistic insight from real-time analysis of oxidative phosphorylation. PLoS ONE 2018, 13, e0199505. [Google Scholar] [CrossRef] [Green Version]
- Bhatti, J.S.; Bhatti, G.K.; Reddy, P.H. Mitochondrial dysfunction and oxidative stress in metabolic disorders—A step towards mitochondria based therapeutic strategies. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1066–1077. [Google Scholar] [CrossRef]
- Rimessi, A.; Previati, M.; Nigro, F.; Wieckowski, M.R.; Pinton, P. Mitochondrial reactive oxygen species and inflammation: Molecular mechanisms, diseases and promising therapies. Int. J. Biochem. Cell Biol. 2016, 81, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Garaude, J.; Acín-Pérez, R.; Martínez-Cano, S.; Enamorado, M.; Ugolini, M.; Nistal-Villán, E.; Hervás-Stubbs, S.; Pelegrín, P.; Sander, L.E.; Enríquez, J.A.; et al. Mitochondrial respiratory-chain adaptations in macrophages contribute to antibacterial host defense. Nat. Immunol. 2016, 17, 1037–1045. [Google Scholar] [CrossRef] [Green Version]
- Chistiakov, D.A.; Sobenin, I.A.; Revin, V.V.; Orekhov, A.N.; Bobryshev, Y.V. Mitochondrial Aging and Age-Related Dysfunction of Mitochondria. BioMed. Res. Int. 2014, 2014, 238463. [Google Scholar] [CrossRef] [Green Version]
- Bauer, M.E.; De la Fuente, M. The role of oxidative and inflammatory stress and persistent viral infections in immunosenescence. Mech. Ageing Dev. 2016, 158, 27–37. [Google Scholar] [CrossRef]
- Coperchini, F.; Chiovato, L.; Rotondi, M. Interleukin-6, CXCL10 and Infiltrating Macrophages in COVID-19-Related Cytokine Storm: Not One for All But All for One! Front. Immunol. 2021, 12, 668507. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, S.; Howard, S.; Muldoon, O.T.; Whittaker, A.C. Social cohesion and loneliness are associated with the antibody response to COVID-19 vaccination. Brain Behav. Immun. 2022, 103, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-C.; Huang, G.-J.; Lin, J.-G. Chinese herbal prescriptions for COVID-19 management: Special reference to Taiwan Chingguan Yihau (NRICM101). Front. Pharmacol. 2022, 13. [Google Scholar] [CrossRef]
- Piras, S.; Furfaro, A.L.; Domenicotti, C.; Traverso, N.; Marinari, U.M.; Pronzato, M.A.; Nitti, M. RAGE Expression and ROS Generation in Neurons: Differentiation versus Damage. Oxidative Med. Cell. Longev. 2016, 2016, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ottum, M.S.; Mistry, A.M. Advanced glycation end-products: Modifiable environmental factors profoundly mediate insulin resistance. J. Clin. Biochem. Nutr. 2015, 57, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Walker, D.; Lue, L.F.; Paul, G.; Patel, A.; Sabbagh, M.N. Receptor for advanced glycation endproduct modulators: A new therapeutic target in Alzheimer’s disease. Expert Opin. Investig. Drugs 2014, 24, 393–399. [Google Scholar] [CrossRef] [Green Version]
- Gebhardt, C.; Riehl, A.; Durchdewald, M.; Németh, J.; Fürstenberger, G.; Müller-Decker, K.; Enk, A.; Arnold, B.; Bierhaus, A.; Nawroth, P.P.; et al. RAGE signaling sustains inflammation and promotes tumor development. J. Exp. Med. 2008, 205, 275–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparvero, L.J.; Asafu-Adjei, D.; Kang, R.; Tang, D.; Amin, N.; Im, J.; Rutledge, R.; Lin, B.; Amoscato, A.A.; Zeh, H.J.; et al. RAGE (Receptor for Advanced Glycation Endproducts), RAGE Ligands, and their role in Cancer and Inflammation. J. Transl. Med. 2009, 7, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, M.S.; Fotheringham, A.K.; Cooper, M.E.; Forbes, J.M. Targeting advanced glycation endproducts and mitochondrial dysfunction in cardiovascular disease. Curr. Opin. Pharmacol. 2013, 13, 654–661. [Google Scholar] [CrossRef] [PubMed]











Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrade, F.d.O.; Jin, L.; Clarke, R.; Wood, I.; Dutton, M.; Anjorin, C.; Rubin, G.; Gao, A.; Sengupta, S.; FitzGerald, K.; et al. Social Isolation Activates Dormant Mammary Tumors, and Modifies Inflammatory and Mitochondrial Metabolic Pathways in the Rat Mammary Gland. Cells 2023, 12, 961. https://doi.org/10.3390/cells12060961
Andrade FdO, Jin L, Clarke R, Wood I, Dutton M, Anjorin C, Rubin G, Gao A, Sengupta S, FitzGerald K, et al. Social Isolation Activates Dormant Mammary Tumors, and Modifies Inflammatory and Mitochondrial Metabolic Pathways in the Rat Mammary Gland. Cells. 2023; 12(6):961. https://doi.org/10.3390/cells12060961
Chicago/Turabian StyleAndrade, Fabia de Oliveira, Lu Jin, Robert Clarke, Imani Wood, MaryAnn Dutton, Chezaray Anjorin, Grace Rubin, Audrey Gao, Surojeet Sengupta, Kevin FitzGerald, and et al. 2023. "Social Isolation Activates Dormant Mammary Tumors, and Modifies Inflammatory and Mitochondrial Metabolic Pathways in the Rat Mammary Gland" Cells 12, no. 6: 961. https://doi.org/10.3390/cells12060961
APA StyleAndrade, F. d. O., Jin, L., Clarke, R., Wood, I., Dutton, M., Anjorin, C., Rubin, G., Gao, A., Sengupta, S., FitzGerald, K., & Hilakivi-Clarke, L. (2023). Social Isolation Activates Dormant Mammary Tumors, and Modifies Inflammatory and Mitochondrial Metabolic Pathways in the Rat Mammary Gland. Cells, 12(6), 961. https://doi.org/10.3390/cells12060961