High Intrinsic Oncogenic Potential in the Myc-Box-Deficient Hydra Myc3 Protein
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Whole Mount In Situ Hybridization
2.3. Single-Cell Transcriptome Analyses
2.4. Cells and Retroviruses
2.5. Protein Expression and Purification
2.6. Protein Analysis
2.7. Protein–DNA Interaction Analysis
2.8. Protein Structure and Homology Modeling
3. Results
3.1. Structure of the Hydra myc3 Gene and Its Protein Product
3.2. Expression of the Hydra myc3 mRNA
3.3. Biochemical Properties of the Hydra Myc3 Protein
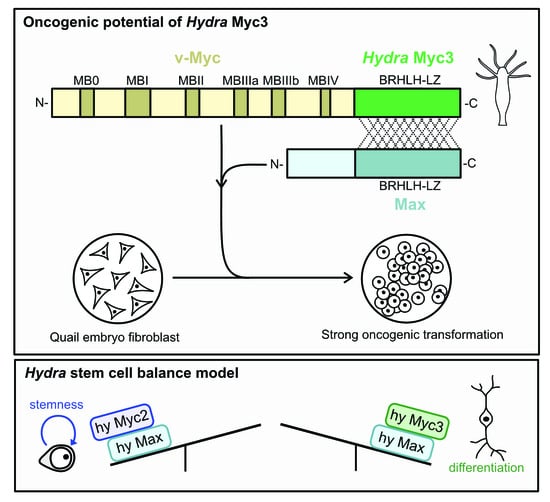
3.4. Oncogenic Potential of the Hydra Myc3 Protein
3.5. Homology Modeling of Hydra Myc3 Predicts a High Stability of Myc3/Max Heterodimers
3.6. Conservation of Critical Amino Acid Residues in the Hydra Myc3 Leucine Zipper Region
4. Discussion
4.1. Myc/Max Dimerization and Oncogenic Transformation
4.2. A Possible Role for Hydra Myc3 in Balancing Stemness and Differentiation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Duesberg, P.H.; Bister, K.; Vogt, P.K. The RNA of avian acute leukemia virus MC29. Proc. Natl. Acad. Sci. USA 1977, 74, 4320–4324. [Google Scholar] [CrossRef] [PubMed]
- Bister, K.; Hayman, M.J.; Vogt, P.K. Defectiveness of avian myelocytomatosis virus MC29: Isolation of long-term nonproducer cultures and analysis of virus-specific polypeptide synthesis. Virology 1977, 82, 431–448. [Google Scholar] [CrossRef]
- Stefan, E.; Bister, K. MYC and RAF: Key effectors in cellular signaling and major drivers in human cancer. Curr. Top. Microbiol. Immunol. 2017, 407, 117–151. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, S.; Eilers, M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell. Biol. 2005, 6, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Conacci-Sorrell, M.; McFerrin, L.; Eisenman, R.N. An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 2014, 4, a014357. [Google Scholar] [CrossRef]
- Vennstrom, B.; Sheiness, D.; Zabielski, J.; Bishop, J.M. Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J. Virol. 1982, 42, 773–779. [Google Scholar] [CrossRef]
- Eisenman, R.N. Deconstructing myc. Genes. Dev. 2001, 15, 2023–2030. [Google Scholar] [CrossRef]
- Kalkat, M.; De Melo, J.; Hickman, K.A.; Lourenco, C.; Redel, C.; Resetca, D.; Tamachi, A.; Tu, W.B.; Penn, L.Z. MYC deregulation in primary human cancers. Genes 2017, 8, 151. [Google Scholar] [CrossRef]
- Nesbit, C.E.; Tersak, J.M.; Prochownik, E.V. MYC oncogenes and human neoplastic disease. Oncogene 1999, 18, 3004–3016. [Google Scholar] [CrossRef]
- Baluapuri, A.; Wolf, E.; Eilers, M. Target gene-independent functions of MYC oncoproteins. Nat. Rev. Mol. Cell. Biol. 2020, 21, 255–267. [Google Scholar] [CrossRef]
- Herold, S.; Herkert, B.; Eilers, M. Facilitating replication under stress: An oncogenic function of MYC? Nat. Rev. Cancer 2009, 9, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Blackwood, E.M.; Eisenman, R.N. Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 1991, 251, 1211–1217. [Google Scholar] [CrossRef]
- Wolf, E.; Lin, C.Y.; Eilers, M.; Levens, D.L. Taming of the beast: Shaping Myc-dependent amplification. Trends Cell Biol. 2015, 25, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 1999, 19, 1–11. [Google Scholar] [CrossRef]
- Gallant, P.; Shiio, Y.; Cheng, P.F.; Parkhurst, S.M.; Eisenman, R.N. Myc and Max homologs in Drosophila. Science 1996, 274, 1523–1527. [Google Scholar] [CrossRef] [PubMed]
- Orian, A.; van Steensel, B.; Delrow, J.; Bussemaker, H.J.; Li, L.; Sawado, T.; Williams, E.; Loo, L.W.; Cowley, S.M.; Yost, C.; et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003, 17, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Hartl, M.; Mitterstiller, A.M.; Valovka, T.; Breuker, K.; Hobmayer, B.; Bister, K. Stem cell-specific activation of an ancestral myc protooncogene with conserved basic functions in the early metazoan Hydra. Proc. Natl. Acad. Sci. USA 2010, 107, 4051–4056. [Google Scholar] [CrossRef]
- Young, S.L.; Diolaiti, D.; Conacci-Sorrell, M.; Ruiz-Trillo, I.; Eisenman, R.N.; King, N. Premetazoan ancestry of the Myc-Max network. Mol. Biol. Evol. 2011, 28, 2961–2971. [Google Scholar] [CrossRef]
- Hobmayer, B.; Jenewein, M.; Eder, D.; Eder, M.K.; Glasauer, S.; Gufler, S.; Hartl, M.; Salvenmoser, W. Stemness in Hydra-a current perspective. Int. J. Dev. Biol. 2012, 56, 509–517. [Google Scholar] [CrossRef]
- Vogg, M.C.; Galliot, B.; Tsiairis, C.D. Model systems for regeneration: Hydra. Development 2019, 146, dev177212. [Google Scholar] [CrossRef] [PubMed]
- Gierer, A.; Meinhardt, H. A theory of biological pattern formation. Kybernetik 1972, 12, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.R. Axial patterning in hydra. Cold Spring Harb. Perspect. Biol. 2009, 1, a000463. [Google Scholar] [CrossRef] [PubMed]
- Bosch, T.C.; Anton-Erxleben, F.; Hemmrich, G.; Khalturin, K. The Hydra polyp: Nothing but an active stem cell community. Dev. Growth Differ. 2010, 52, 15–25. [Google Scholar] [CrossRef]
- Hartl, M.; Glasauer, S.; Valovka, T.; Breuker, K.; Hobmayer, B.; Bister, K. Hydra myc2, a unique pre-bilaterian member of the myc gene family, is activated in cell proliferation and gametogenesis. Biol. Open 2014, 3, 397–407. [Google Scholar] [CrossRef]
- Hobmayer, B.; Holstein, T.W.; David, C.N. Stimulation of tentacle and bud formation by the neuropeptide head activator in Hydra magnipapillata. Dev. Biol. 1997, 183, 1–8. [Google Scholar] [CrossRef]
- Philipp, I.; Holstein, T.W.; Hobmayer, B. HvJNK, a Hydra member of the c-Jun NH2-terminal kinase gene family, is expressed during nematocyte differentiation. Gene Expr. Patterns 2005, 5, 397–402. [Google Scholar] [CrossRef]
- Cazet, J.F.; Siebert, S.; Morris Little, H.; Bertemes, P.; Primack, A.S.; Ladurner, P.; Achrainer, M.; Fredriksen, M.T.; Moreland, R.T.; Singh, S.; et al. A chromosome-scale epigenetic map of the Hydra genome reveals conserved regulators of cell state. Genome Res. 2023, 33, 283–298. [Google Scholar] [CrossRef]
- Siebert, S.; Farrell, J.A.; Cazet, J.F.; Abeykoon, Y.; Primack, A.S.; Schnitzler, C.E.; Juliano, C.E. Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 2019, 365, aav9314. [Google Scholar] [CrossRef]
- Chari, T.; Weissbourd, B.; Gehring, J.; Ferraioli, A.; Leclere, L.; Herl, M.; Gao, F.; Chevalier, S.; Copley, R.R.; Houliston, E.; et al. Whole-animal multiplexed single-cell RNA-seq reveals transcriptional shifts across Clytia medusa cell types. Sci. Adv. 2021, 7, eabh1683. [Google Scholar] [CrossRef]
- Raffeiner, P.; Schraffl, A.; Schwarz, T.; Röck, R.; Ledolter, K.; Hartl, M.; Konrat, R.; Stefan, E.; Bister, K. Calcium-dependent binding of Myc to calmodulin. Oncotarget 2017, 8, 3327–3343. [Google Scholar] [CrossRef] [PubMed]
- Gufler, S.; Artes, B.; Bielen, H.; Krainer, I.; Eder, M.K.; Falschlunger, J.; Bollmann, A.; Ostermann, T.; Valovka, T.; Hartl, M.; et al. beta-Catenin acts in a position-independent regeneration response in the simple eumetazoan Hydra. Dev. Biol. 2018, 433, 310–323. [Google Scholar] [CrossRef] [PubMed]
- Hartl, M.; Puglisi, K.; Nist, A.; Raffeiner, P.; Bister, K. The brain acid-soluble protein 1 (BASP1) interferes with the oncogenic capacity of MYC and its binding to calmodulin. Mol. Oncol. 2020, 14, 625–644. [Google Scholar] [CrossRef] [PubMed]
- Hartl, M.; Glasauer, S.; Gufler, S.; Raffeiner, A.; Puglisi, K.; Breuker, K.; Bister, K.; Hobmayer, B. Differential regulation of myc homologs by Wnt/beta-Catenin signaling in the early metazoan Hydra. FEBS J. 2019, 286, 2295–2310. [Google Scholar] [CrossRef]
- Nair, S.K.; Burley, S.K. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 2003, 112, 193–205. [Google Scholar] [CrossRef]
- David, C.N.; Gierer, A. Cell cycle kinetics and development of Hydra attenuata. III. Nerve and nematocyte differentiation. J. Cell Sci. 1974, 16, 359–375. [Google Scholar] [CrossRef]
- Heimfeld, S.; Bode, H.R. Growth regulation of the interstitial cell population in hydra. IV. Control of nerve cell and nematocyte differentiation by amplification of non-stem interstitial cells. Dev. Biol. 1986, 116, 59–68. [Google Scholar] [CrossRef]
- Hager, G.; David, C.N. Pattern of differentiated nerve cells in hydra is determined by precursor migration. Development 1997, 124, 569–576. [Google Scholar] [CrossRef]
- Fieber, W.; Schneider, M.L.; Matt, T.; Krautler, B.; Konrat, R.; Bister, K. Structure, function, and dynamics of the dimerization and DNA-binding domain of oncogenic transcription factor v-Myc. J. Mol. Biol. 2001, 307, 1395–1410. [Google Scholar] [CrossRef]
- Masso-Valles, D.; Soucek, L. Blocking Myc to Treat Cancer: Reflecting on Two Decades of Omomyc. Cells 2020, 9, 883. [Google Scholar] [CrossRef]
- Annibali, D.; Whitfield, J.R.; Favuzzi, E.; Jauset, T.; Serrano, E.; Cuartas, I.; Redondo-Campos, S.; Folch, G.; Gonzalez-Junca, A.; Sodir, N.M.; et al. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat. Commun. 2014, 5, 4632. [Google Scholar] [CrossRef] [PubMed]
- Eilers, M.; Eisenman, R.N. Myc’s broad reach. Genes. Dev. 2008, 22, 2755–2766. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. Enigmatic MYC Conducts an Unfolding Systems Biology Symphony. Genes. Cancer 2010, 1, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Fagnocchi, L.; Cherubini, A.; Hatsuda, H.; Fasciani, A.; Mazzoleni, S.; Poli, V.; Berno, V.; Rossi, R.L.; Reinbold, R.; Endele, M.; et al. A Myc-driven self-reinforcing regulatory network maintains mouse embryonic stem cell identity. Nat. Commun. 2016, 7, 11903. [Google Scholar] [CrossRef] [PubMed]
- Hartl, M.; Bister, K. MYC Analysis in Cancer and Evolution. Methods Mol. Biol. 2021, 2318, 87–117. [Google Scholar] [CrossRef] [PubMed]
- Hobmayer, B.; Rentzsch, F.; Kuhn, K.; Happel, C.M.; von Laue, C.C.; Snyder, P.; Rothbacher, U.; Holstein, T.W. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 2000, 407, 186–189. [Google Scholar] [CrossRef]
- Holstein, T.W. The role of cnidarian developmental biology in unraveling axis formation and Wnt signaling. Dev. Biol. 2022, 487, 74–98. [Google Scholar] [CrossRef]
- Holstein, T.W.; Hobmayer, E.; Technau, U. Cnidarians: An evolutionarily conserved model system for regeneration? Dev. Dyn. 2003, 226, 257–267. [Google Scholar] [CrossRef]
- Chapman, J.A.; Kirkness, E.F.; Simakov, O.; Hampson, S.E.; Mitros, T.; Weinmaier, T.; Rattei, T.; Balasubramanian, P.G.; Borman, J.; Busam, D.; et al. The dynamic genome of Hydra. Nature 2010, 464, 592–596. [Google Scholar] [CrossRef]
- Klimovich, A.; Wittlieb, J.; Bosch, T.C.G. Transgenesis in Hydra to characterize gene function and visualize cell behavior. Nat. Protoc. 2019, 14, 2069–2090. [Google Scholar] [CrossRef]
- Vogg, M.C.; Galliot, B. Combining RNAi-Mediated beta-Catenin Inhibition and Reaggregation to Study Hydra Whole-Body Regeneration. Methods Mol. Biol. 2022, 2450, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Ambrosone, A.; Marchesano, V.; Tino, A.; Hobmayer, B.; Tortiglione, C. Hymyc1 downregulation promotes stem cell proliferation in Hydra vulgaris. PLoS ONE 2012, 7, e30660. [Google Scholar] [CrossRef] [PubMed]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Castell, A.; Yan, Q.; Fawkner, K.; Hydbring, P.; Zhang, F.; Verschut, V.; Franco, M.; Zakaria, S.M.; Bazzar, W.; Goodwin, J.; et al. A selective high affinity MYC-binding compound inhibits MYC:MAX interaction and MYC-dependent tumor cell proliferation. Sci. Rep. 2018, 8, 10064. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, M.E.; Jauset, T.; Masso-Valles, D.; Martinez-Martin, S.; Rahl, P.; Maltais, L.; Zacarias-Fluck, M.F.; Casacuberta-Serra, S.; Serrano Del Pozo, E.; Fiore, C.; et al. Intrinsic cell-penetrating activity propels Omomyc from proof of concept to viable anti-MYC therapy. Sci. Transl. Med. 2019, 11, eaar5012. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhang, J.; Wang, Y.; Liu, L.; Gao, J. Computational insights into the differentiated binding affinities of Myc, Max, and Omomyc dimers to the E-boxes of DNA. J. Mol. Model. 2022, 28, 329. [Google Scholar] [CrossRef]
- Mullard, A. Climbing cancer’s MYC mountain. Nat. Rev. Drug. Discov. 2022, 21, 865–867. [Google Scholar] [CrossRef]
- Khalturin, K.; Anton-Erxleben, F.; Milde, S.; Plotz, C.; Wittlieb, J.; Hemmrich, G.; Bosch, T.C. Transgenic stem cells in Hydra reveal an early evolutionary origin for key elements controlling self-renewal and differentiation. Dev. Biol. 2007, 309, 32–44. [Google Scholar] [CrossRef]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Berking, S. Commitment of stem cells to nerve cells and migration of nerve cells precursors in preparatory bud development in Hydra. J. Embryol. Exp. Morphol. 1980, 60, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Otto, J.J.; Campbell, R.D. Budding in Hydra attenuata: Bud stages and fate map. J. Exp. Zool. 1977, 200, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Soucek, L.; Whitfield, J.; Martins, C.P.; Finch, A.J.; Murphy, D.J.; Sodir, N.M.; Karnezis, A.N.; Swigart, L.B.; Nasi, S. Modelling Myc inhibition as a cancer therapy. Nature 2008, 455, 679–683. [Google Scholar] [CrossRef] [PubMed]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lechable, M.; Tang, X.; Siebert, S.; Feldbacher, A.; Fernández-Quintero, M.L.; Breuker, K.; Juliano, C.E.; Liedl, K.R.; Hobmayer, B.; Hartl, M. High Intrinsic Oncogenic Potential in the Myc-Box-Deficient Hydra Myc3 Protein. Cells 2023, 12, 1265. https://doi.org/10.3390/cells12091265
Lechable M, Tang X, Siebert S, Feldbacher A, Fernández-Quintero ML, Breuker K, Juliano CE, Liedl KR, Hobmayer B, Hartl M. High Intrinsic Oncogenic Potential in the Myc-Box-Deficient Hydra Myc3 Protein. Cells. 2023; 12(9):1265. https://doi.org/10.3390/cells12091265
Chicago/Turabian StyleLechable, Marion, Xuechen Tang, Stefan Siebert, Angelika Feldbacher, Monica L. Fernández-Quintero, Kathrin Breuker, Celina E. Juliano, Klaus R. Liedl, Bert Hobmayer, and Markus Hartl. 2023. "High Intrinsic Oncogenic Potential in the Myc-Box-Deficient Hydra Myc3 Protein" Cells 12, no. 9: 1265. https://doi.org/10.3390/cells12091265
APA StyleLechable, M., Tang, X., Siebert, S., Feldbacher, A., Fernández-Quintero, M. L., Breuker, K., Juliano, C. E., Liedl, K. R., Hobmayer, B., & Hartl, M. (2023). High Intrinsic Oncogenic Potential in the Myc-Box-Deficient Hydra Myc3 Protein. Cells, 12(9), 1265. https://doi.org/10.3390/cells12091265