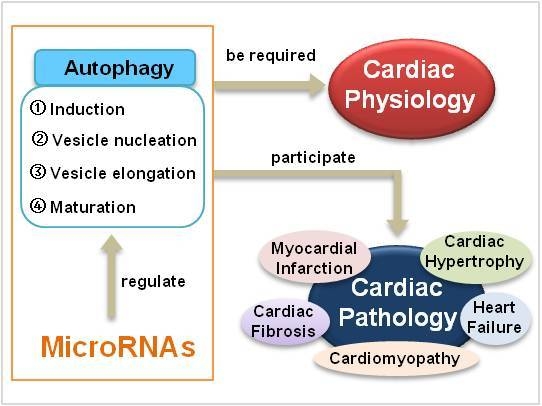
MicroRNAs in Cardiac Autophagy: Small Molecules and Big Role
Abstract
:1. Overview of Autophagy and MicroRNAs
1.1. Autophagy
1.2. MicroRNAs
2. MicroRNAs Regulate the Core Autophagy Signaling Cascades
3. The Role of MicroRNAs in Cardiac Autophagy
4. MicroRNAs in Autophagy-Related Heart Diseases
4.1. Myocardial Infarction
4.2. Cardiac Hypertrophy
4.3. Cardiac Fibrosis
4.4. Cardiomyopathy
4.5. Heart Failure
5. Conclusions and Perspectives
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| ACE2 | angiotensin-converting enzyme 2 |
| AGOs | argonaute proteins |
| AMPK | AMP- activated protein kinase |
| ARC | apoptosis repressor with caspase recruit domain |
| ATG | autophagy-related gene |
| BAG3 | Bcl-2-associated athanogene 3 |
| Bcl2-L-13 | Bcl2-like protein 13 |
| BNIP3 | Bcl-2 interacting protein 3 |
| BNIP3L | Bcl2 interacting protein 3 like |
| CDS | coding sequence |
| CISD2 | CDGSH iron sulfur domain 2 |
| FIP200 | focal adhesion kinase family interacting protein of 200 kDa |
| FOXO3 | forkhead box O3 |
| FUNDC1 | FUN14 domain containing protein 13 |
| GSK3β | glycogen synthase kinase 3β |
| HCM | hypertrophic cardiomyopathy |
| MFN2 | mitofusion2 |
| miRNAs | microRNAs |
| mTOR | mammalian target of rapamycin |
| mTORC1 | mTOR complex 1 |
| PARP-1 | poly (ADP-ribose) polymerase 1 |
| PE | phosphatidylethanolamine |
| PIK3C3 | phosphatidylinositol 3-kinase catalytic subunit type 3 |
| PINK1 | putative kinase 1 |
| PTEN | phosphatase and tensin homolog deleted on chromosome ten |
| TSC1 | tuberous sclerosis complex 1 |
| TTC | Takatsubo cardiomyopathy |
| ULK | Unc-51-like kinase |
| UTR | untranslated region |
| UVRAG | UV irradiation resistance-associated gene |
References
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Legakis, J.E.; Yen, W.L.; Klionsky, D.J. A cycling protein complex required for selective autophagy. Autophagy 2007, 3, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Kiriyama, Y.; Nochi, H. Intra- and intercellular quality control mechanisms of mitochondria. Cells 2017, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.W.; Ordureau, A.; Heo, J.M. Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 93–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, R.; Pattison, J.S. Macroautophagy and chaperone-mediated autophagy in heart failure: The known and the unknown. Oxid. Med. Cell. Longev. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dai, C.; Fan, Y.; Guo, B.; Ren, K.; Sun, T.; Wang, W. From autophagy to mitophagy: The roles of p62 in neurodegenerative diseases. J. Bioenerg. Biomembr. 2017, 49, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Li, T.S. Dual role of mitophagy in cancer drug resistance. Anticancer Res. 2018, 38, 617–621. [Google Scholar] [PubMed]
- Herst, P.M.; Rowe, M.R.; Carson, G.M.; Berridge, M.V. Functional mitochondria in health and disease. Front. Endocrinol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Cummins, N.; Gotz, J. Shedding light on mitophagy in neurons: What is the evidence for pink1/parkin mitophagy in vivo? Cell. Mol. Life Sci. 2018, 75, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, L.; Xue, Y.; Ma, Y.; Liu, X.; Li, Z.; Li, Z.; Liu, Y. Endothelial monocyte-activating polypeptide-ii induces bnip3-mediated mitophagy to enhance temozolomide cytotoxicity of glioma stem cells via down-regulating mir-24-3p. Front. Mol. Neurosci. 2018, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Murakawa, T.; Yamaguchi, O.; Hashimoto, A.; Hikoso, S.; Takeda, T.; Oka, T.; Yasui, H.; Ueda, H.; Akazawa, Y.; Nakayama, H.; et al. Bcl-2-like protein 13 is a mammalian atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun. 2015, 6, 7527. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhu, P.; Wang, J.; Zhu, H.; Ren, J.; Chen, Y. Pathogenesis of cardiac ischemia reperfusion injury is associated with ck2alpha-disturbed mitochondrial homeostasis via suppression of fundc1-related mitophagy. Cell Death Differ. 2018, 25, 1080. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Zhang, Y.H.; Li, R.B.; Zhou, L.Y.; An, T.; Zhang, R.C.; Zhai, M.; Huang, Y.; Yan, K.W.; Dong, Y.H.; et al. Lncrna caif inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat. Commun. 2018, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Song, Y.; Liu, L.; Hou, N.; An, X.; Zhan, D.; Li, Y. Mir-199a impairs autophagy and induces cardiac hypertrophy through mtor activation. Cell Death Differ. 2017, 24, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, C.; Turdi, S.; Richmond, K.L.; Zhang, Y.; Ren, J. ALDH2 protects against high fat diet-induced obesity cardiomyopathy and defective autophagy: Role of CaM kinase II, histone H3K9 methyltransferase SUV39H, Sirt1, and PGC-1α deacetylation. Int. J. Obes. 2018, 42, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Wang, F.; Gao, R.; Wu, J.; Ou, Y.; Chen, X.; Wang, T.; Zhou, X.; Zhu, W.; Li, P.; et al. Autophagy inhibition of hsa-miR-19a-3p/19b-3p by targeting TGF-β R II during TGF-β1-induced fibrogenesis in human cardiac fibroblasts. Sci. Rep. 2016, 6, 24747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, M.; Wang, J.; Wang, C.; Wang, X.; Dong, W.; Qiu, W.; Wang, Y.; Zhao, X.; Zou, Y.; Song, L.; et al. MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell Death Differ. 2015, 22, 986–999. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, C.Y.; Zhou, L.Y.; Wang, J.X.; Wang, M.; Zhao, B.; Zhao, W.K.; Xu, S.J.; Fan, L.H.; Zhang, X.J.; et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 2015, 6, 6779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higashi, K.; Yamada, Y.; Minatoguchi, S.; Baba, S.; Iwasa, M.; Kanamori, H.; Kawasaki, M.; Nishigaki, K.; Takemura, G.; Kumazaki, M.; et al. MicroRNA-145 repairs infarcted myocardium by accelerating cardiomyocyte autophagy. Am. J. Physiology-Heart Circ. Physiol. 2015, 309, H1813–H1826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ucar, A.; Gupta, S.K.; Fiedler, J.; Erikci, E.; Kardasinski, M.; Batkai, S.; Dangwal, S.; Kumarswamy, R.; Bang, C.; Holzmann, A.; et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat. Commun. 2012, 3, 1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Li, Y.; Wang, Y.; Qian, J.; Ma, H.; Wang, X.; Jiang, G.; Liu, M.; An, Y.; Ma, L.; et al. Cardiomyocyte-restricted low density lipoprotein receptor-related protein 6 (LRP6) deletion leads to lethal dilated cardiomyopathy partly through Drp1 signaling. Theranostics 2018, 8, 627–643. [Google Scholar] [CrossRef] [PubMed]
- Carresi, C.; Musolino, V.; Gliozzi, M.; Maiuolo, J.; Mollace, R.; Nucera, S.; Maretta, A.; Sergi, D.; Muscoli, S.; Gratteri, S.; et al. Anti-oxidant effect of bergamot polyphenolic fraction counteracts doxorubicin-induced cardiomyopathy: Role of autophagy and c-kitposCD45negCD31neg cardiac stem cell activation. J. Mol. Cell. Cardiol. 2018, 119, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.H.; Nam, J.W.; Heo, I.; Rhee, J.K.; Sohn, S.Y.; Cho, Y.; Zhang, B.T.; Kim, V.N. Molecular basis for the recognition of primary micrornas by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Hutvagner, G.; McLachlan, J.; Pasquinelli, A.E.; Balint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Huntzinger, E.; Izaurralde, E. Getting to the root of miRNA-mediated gene silencing. Cell 2008, 132, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Yang, Z.; Xu, Y.; Chen, Y.; Yu, Q. Micrornas in apoptosis, autophagy and necroptosis. Oncotarget 2015, 6, 8474–8490. [Google Scholar] [CrossRef] [PubMed]
- Llorens, F.; Thune, K.; Marti, E. Regional and subtype-dependent miRNA signatures in sporadic Creutzfeldt-Jakob disease are accompanied by alterations in miRNA silencing machinery and biogenesis. PloS Pathog. 2018, 14, e1006802. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ma, B.; Han, X. The role of autophagy in angiotensin II induced pathological cardiac hypertrophy. J. Mol. Endocrinol. 2016, 57, R143–R152. [Google Scholar] [CrossRef] [PubMed]
- Frankel, L.B.; Lund, A.H. Microrna regulation of autophagy. Carcinogenesis 2012, 33, 2018–2025. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Z.; Lu, Y.; Song, K.; Liu, X.; Xia, F.; Sun, W. Downregulation of UKL1 by microRNA-372 inhibits the survival of human pancreatic adenocarcinoma cells. Cancer Sci. 2017, 108, 1811–1819. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Zhang, T.; Ding, S.; Wei, J.; Su, C.; Liu, H.; Xu, G. MicroRNA-17-5p modulates bacille calmette-guerin growth in RAW264.7 cells by targeting ULK1. PloS ONE 2015, 10, e0138011. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, S.I.; Gautschi, O.; Batliner, J.; Gugger, M.; Fey, M.F.; Tschan, M.P. MicroRNA-106a targets autophagy and enhances sensitivity of lung cancer cells to Src inhibitors. Lung Cancer 2017, 107, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chuang, A.Y.; Ratovitski, E.A. Phospho-ΔNp63α/miR-885–3p axis in tumor cell life and cell death upon cisplatin exposure. Cell Cycle 2011, 10, 3938–3947. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, J.; Tang, N. Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience 2017, 354, 1–10. [Google Scholar] [CrossRef] [PubMed]
- John Clotaire, D.Z.; Zhang, B.; Wei, N.; Gao, R.; Zhao, F.; Wang, Y.; Lei, M.; Huang, W. Mir-26b inhibits autophagy by targeting ULK2 in prostate cancer cells. Biochem. Biophys. Res. Commun. 2016, 472, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Qiang, Q.; Shan, H.; Shi, M.; Gan, G.; Ma, F.; Chen, B. MiR-20a and miR-20b negatively regulate autophagy by targeting RB1CC1/FIP200 in breast cancer cells. Life Sci. 2016, 147, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Ren, X.; Hait, W.N.; Yang, J.M. Therapeutic targeting of autophagy in disease: Biology and pharmacology. Pharmacol. Rev. 2013, 65, 1162–1197. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guerrero-Preston, R.; Ratovitski, E.A. Phospho-ΔNp63α-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle 2012, 11, 1247–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Yu, Y.; Li, S.; Liu, Y.; Zhou, S.; Cao, S.; Yin, J.; Li, G. Micro RNA-30a ameliorates hepatic fibrosis by inhibiting Beclin1-mediated autophagy. J. Cell. Mol. Med. 2017, 21, 3679–3692. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Song, L.; Zhao, Y.; Liu, Q.; Zhang, S. Inhibition of Beclin-1-mediated autophagy by microRNA-17-5p enhanced the radiosensitivity of glioma cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Zhang, Y.; Kang, L.; Song, Y.; Wang, K.; Li, S.; Wu, X.; Hua, W.; Shao, Z.; Yang, S.; et al. Methylation of microRNA-129-5p modulates nucleus pulposus cell autophagy by targeting Beclin-1 in intervertebral disc degeneration. Oncotarget 2017, 8, 86264–86276. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shi, H.; Lin, S.; Ba, M.; Cui, S. MicroRNA-216a enhances the radiosensitivity of pancreatic cancer cells by inhibiting beclin-1-mediated autophagy. Oncol. Rep. 2015, 34, 1557–1564. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, G.; le Sage, C.; Tekirdag, K.A.; Agami, R.; Gozuacik, D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy 2012, 8, 165–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, A.G.; Meng, F.G.; Wang, M.G. Cisd2 promotes the proliferation of glioma cells via suppressing beclin1mediated autophagy and is targeted by microRNA449a. Mol. Med. Rep. 2017, 16, 7939–7948. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Ji, X.; Rong, R.; Li, Y.; Yao, W.; Yuan, J.; Wu, Q.; Yang, J.; Yan, W.; Han, L.; et al. MiR-449a regulates autophagy to inhibit silica-induced pulmonary fibrosis through targeting Bcl2. J. Mol. Med. 2016, 94, 1267–1279. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Gao, X.; Wang, J.; Yang, C.; Wang, Y.; Liu, Y.; Zou, W.; Liu, T. Hypoxia-induced microRNA-146a represses Bcl-2 through Traf6/IRAK1 but not Smad4 to promote chondrocyte autophagy. Biol. Chem. 2017, 398, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.L.; He, G.Y.; Lan, X.L.; Zeng, Z.C.; Guan, J.; Ding, Y.; Qian, X.L.; Liao, W.T.; Ding, Y.Q.; Liang, L. Inhibition of ATG12-mediated autophagy by miR-214 enhances radiosensitivity in colorectal cancer. Oncogenesis 2018, 7, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, D.; Zhou, C.; Han, S.; Hou, X.; Kang, S.; Zhang, Y. MicroRNA-378 enhances migration and invasion in cervical cancer by directly targeting autophagy-related protein 12. Mol. Med. Rep. 2018, 17, 6319–6326. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Yu, B.; Cheng, C.; Cheng, T.; Yuan, B.; Li, K.; Xiao, J.; Qiu, Z. Mir505-3p regulates axonal development via inhibiting the autophagy pathway by targeting Atg12. Autophagy 2017, 13, 1679–1696. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Liu, A.; Zhang, J.; Ji, W.; Li, Y.; Yang, X.; Wu, Z.; Guo, J. miR-23b improves cognitive impairments in traumatic brain injury by targeting ATG12-mediated neuronal autophagy. Behav. Brain Res. 2018, 340, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; He, Y.; Zhai, N.; Ding, S.; Li, J.; Peng, Z. MicroRNA-181a inhibits autophagy by targeting Atg5 in hepatocellular carcinoma. Front. Biosci. (Landmark edition) 2018, 23, 388–396. [Google Scholar]
- Fu, X.T.; Shi, Y.H.; Zhou, J.; Peng, Y.F.; Liu, W.R.; Shi, G.M.; Gao, Q.; Wang, X.Y.; Song, K.; Fan, J.; et al. MicroRNA-30a suppresses autophagy-mediated anoikis resistance and metastasis in hepatocellular carcinoma. Cancer Lett. 2018, 412, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Chen, J.; Zhou, H.; Chen, Y.; Zhi, Y.; Zhang, B.; Chen, L.; Chu, X.; Wang, R.; Zhang, C. Pu.1/microRNA-142-3p targets ATG5/ATG16L1 to inactivate autophagy and sensitize hepatocellular carcinoma cells to sorafenib. Cell Death Dis. 2018, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Gan, J.; Cai, Q.; Qu, Y.; Zhao, F.; Wan, C.; Luo, R.; Mu, D. MiR-96 attenuates status epilepticus-induced brain injury by directly targeting Atg7 and Atg16l1. Sci. Rep. 2017, 7, 10270. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, Z.Z.; Yang, W.; Ouyang, Z.H.; Xue, J.B.; Li, X.L.; Zhang, J.; Chen, W.K.; Yan, Y.G.; Wang, W.J. MiR-210 facilitates ECM degradation by suppressing autophagy via silencing of ATG7 in human degenerated NP cells. Biomed. Pharmacother. 2017, 93, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Comincini, S.; Allavena, G.; Palumbo, S.; Morini, M.; Durando, F.; Angeletti, F.; Pirtoli, L.; Miracco, C. MicroRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol. Ther. 2013, 14, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shen, W.; Zhu, Z.; Lin, J.; Fang, Q.; Ruan, Y.; Zhao, H. Combined inhibition of EGFR and c-ABL suppresses the growth of fulvestrant-resistant breast cancer cells through miR-375-autophagy axis. Biochem. Bioph. Res. Commun. 2018, 498, 559–565. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Cheng, Y. Inhibition of miR-20 promotes proliferation and autophagy in articular chondrocytes by PI3K/AKT/mTOR signaling pathway. Biomed. Pharmacother. 2018, 97, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Dai, X.; Ni, Z.; Yan, X.; He, F.; Lian, J. The downregulation of ATG4B mediated by microRNA-34a/34c-5p suppresses rapamycin-induced autophagy. Irani. J. Basic Med. Sci. 2017, 20, 1125–1130. [Google Scholar]
- Frankel, L.B.; Wen, J.; Lees, M.; Hoyer-Hansen, M.; Farkas, T.; Krogh, A.; Jaattela, M.; Lund, A.H. MicroRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011, 30, 4628–4641. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhu, X.; He, B.; Zhang, Y.; Kang, B.; Wang, Z.; Ni, X. MiR-204 regulates cardiomyocyte autophagy induced by ischemia-reperfusion through LC3-II. J. Biomed. Sci. 2011, 18, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, Q.; Liang, L.; Guo, W.; Zha, R.; Tian, Q.; Huang, S.; Yao, J.; Ding, J.; Bao, M.; Ge, C.; et al. Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology 2011, 54, 2064–2075. [Google Scholar] [CrossRef] [PubMed]
- Rotter, D.; Rothermel, B.A. Targets, trafficking, and timing of cardiac autophagy. Pharmacol. Res. 2012, 66, 494–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.V.; Rothermel, B.A.; Hill, J.A. Autophagy in hypertensive heart disease. J. Biol. Chem. 2010, 285, 8509–8514. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, D.; He, Y.; Melendez, A.; Feng, Z.; Hong, Q.; Bai, X.; Li, Q.; Cai, G.; Wang, J.; et al. MiR-34 modulates caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age 2013, 35, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Kovaleva, V.; Mora, R.; Park, Y.J.; Plass, C.; Chiramel, A.I.; Bartenschlager, R.; Dohner, H.; Stilgenbauer, S.; Pscherer, A.; Lichter, P.; et al. MircoRNA-130a targets ATG2b and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012, 72, 1763–1772. [Google Scholar] [CrossRef] [PubMed]
- Bravo-San Pedro, J.M.; Kroemer, G.; Galluzzi, L. Autophagy and mitophagy in cardiovascular disease. Circ. Res. 2017, 120, 1812–1824. [Google Scholar] [CrossRef] [PubMed]
- Nakai, A.; Yamaguchi, O.; Takeda, T.; Higuchi, Y.; Hikoso, S.; Taniike, M.; Omiya, S.; Mizote, I.; Matsumura, Y.; Asahi, M.; et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007, 13, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Taneike, M.; Yamaguchi, O.; Nakai, A.; Hikoso, S.; Takeda, T.; Mizote, I.; Oka, T.; Tamai, T.; Oyabu, J.; Murakawa, T.; et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy 2010, 6, 600–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouimet, M.; Ediriweera, H.; Afonso, M.S.; Ramkhelawon, B.; Singaravelu, R.; Liao, X.; Bandler, R.C.; Rahman, K.; Fisher, E.A.; Rayner, K.J.; et al. MicroRNA-33 regulates macrophage autophagy in atherosclerosis. Arterioscleros. Thromb. Vasc. Biol. 2017, 37, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, W.N.; Xu, S.B.; Wu, H.; Dai, B.; Jian, D.D.; Yang, M.; Wu, Y.T.; Feng, Q.; Zhu, J.H.; et al. MicroRNA-214-3p: A link between autophagy and endothelial cell dysfunction in atherosclerosis. Acta Physiol. 2018, 222, e12973. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, Z.; Hu, X.; Wan, T.; Wu, H.; Jiang, W.; Hu, R. Human aortic smooth muscle cell-derived exosomal miR-221/222 inhibits autophagy via a PTEN/Akt signaling pathway in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2016, 479, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, Y.T.; Xu, T.H.; Sun, W.; Tian, B.Y.; Sheng, Z.T.; Sun, L.; Liu, L.L.; Ma, J.F.; Wang, L.N.; et al. MicroRNA-30b regulates high phosphorus level-induced autophagy in vascular smooth muscle cells by targeting BECN1. Cell. Physiol. Biochem. 2017, 42, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, F.; Izzo, V.; Niso-Santano, M.; Vacchelli, E.; Galluzzi, L.; Maiuri, M.C.; Kroemer, G. Regulation of autophagy by stress-responsive transcription factors. Semin. Cancer Biol. 2013, 23, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Ding, F.; Wang, X.; Huang, Q.; Zhang, L.; Bi, C.; Hua, B.; Yuan, Y.; Han, Z.; Jin, M.; et al. By targeting Atg7 microRNA-143 mediates oxidative stress-induced autophagy of c-kit+ mouse cardiac progenitor cells. EBioMedicine 2018, 32, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Dorn, G.W., 2nd. Mitochondrial pruning by Nix and BNip3: An essential function for cardiac-expressed death factors. J. Cardiovasc. Transl. Res. 2010, 3, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Song, M.; Csordas, G.; Kelly, D.P.; Matkovich, S.J.; Dorn, G.W., 2nd. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science 2015, 350, aad2459. [Google Scholar] [CrossRef] [PubMed]
- Andres, A.M.; Hernandez, G.; Lee, P.; Huang, C.; Ratliff, E.P.; Sin, J.; Thornton, C.A.; Damasco, M.V.; Gottlieb, R.A. Mitophagy is required for acute cardioprotection by simvastatin. Antioxid. Redox Signal. 2014, 21, 1960–1973. [Google Scholar] [CrossRef] [PubMed]
- Billia, F.; Hauck, L.; Konecny, F.; Rao, V.; Shen, J.; Mak, T.W. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc. Natl. Acad. Sci. USA 2011, 108, 9572–9577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Zhang, X.; Zhuang, H.; Chen, H.G.; Chen, Y.; Tian, W.; Wu, W.; Li, Y.; Wang, S.; Zhang, L.; et al. MicroRNA-137 is a novel hypoxia-responsive microRNA that inhibits mitophagy via regulation of two mitophagy receptors FUNDC1 and NIX. J. Biol. Chem. 2014, 289, 10691–10701. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Fiesel, F.C.; Belmonte, K.C.; Hudec, R.; Wang, W.X.; Kim, C.; Nelson, P.T.; Springer, W.; Kim, J. MiR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by targeting PTEN-induced putative kinase 1 (PINK1). Mol. Neurodegener. 2016, 11, 55. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Liu, L.; Lao, Y.; Liao, W.; Liao, M.; Luo, X.; Wu, J.; Xie, W.; Zhang, Y.; Xu, N. MicroRNA-181a suppresses parkin-mediated mitophagy and sensitizes neuroblastoma cells to mitochondrial uncoupler-induced apoptosis. Oncotarget 2016, 7, 42274–42287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; Li, T. MicroRNA-410 is involved in mitophagy after cardiac ischemia/reperfusion injury by targeting high-mobility group box 1 protein. J. Cell. Biochem. 2018, 119, 2427–2439. [Google Scholar] [CrossRef] [PubMed]
- Verjans, R.; van Bilsen, M.; Schroen, B. MiRNA deregulation in cardiac aging and associated disorders. Int. Rev. Cell Mol. Biol. 2017, 334, 207–263. [Google Scholar] [PubMed]
- Yan, L.; Gao, S.; Ho, D.; Park, M.; Ge, H.; Wang, C.; Tian, Y.; Lai, L.; De Lorenzo, M.S.; Vatner, D.E.; et al. Calorie restriction can reverse, as well as prevent, aging cardiomyopathy. Age 2013, 35, 2177–2182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egan, D.F.; Shackelford, D.B.; Mihaylova, M.M.; Gelino, S.; Kohnz, R.A.; Mair, W.; Vasquez, D.S.; Joshi, A.; Gwinn, D.M.; Taylor, R.; et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011, 331, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, Y.C.; Fang, C.; Russell, R.C.; Kim, J.H.; Fan, W.; Liu, R.; Zhong, Q.; Guan, K.L. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 2013, 152, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Wang, F.; Hu, S.; Yin, C.; Li, X.; Zhao, S.; Wang, J.; Yan, X. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell. signal. 2012, 24, 2179–2186. [Google Scholar] [CrossRef] [PubMed]
- Wan, G.; Xie, W.; Liu, Z.; Xu, W.; Lao, Y.; Huang, N.; Cui, K.; Liao, M.; He, J.; Jiang, Y.; et al. Hypoxia-induced miR155 is a potent autophagy inducer by targeting multiple players in the mtor pathway. Autophagy 2014, 10, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Martins-Marques, T.; Ribeiro-Rodrigues, T.; Pereira, P.; Codogno, P.; Girao, H. Autophagy and ubiquitination in cardiovascular diseases. DNA Cell Biol. 2015, 34, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Zhong, Y.; Cheng, C.; Liu, B.; Wang, L.; Li, A.; Xiong, L.; Liu, S. MiR-30-regulated autophagy mediates angiotensin II-induced myocardial hypertrophy. PloS ONE 2013, 8, e53950. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yang, S.; Li, H.; Yin, Z.; Fan, J.; Zhao, Y.; Gong, W.; Yan, M.; Wang, D.W. MiR30c is involved in diabetic cardiomyopathy through regulation of cardiac autophagy via BECN1. Mol. Therapy-Nucleic Acids 2017, 7, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Sun, W.; Huang, H.; Ye, J.; Pan, W.; Zhong, Y.; Cheng, C.; You, X.; Liu, B.; Xiong, L.; et al. MiR-34a modulates angiotensin ii-induced myocardial hypertrophy by direct inhibition of atg9a expression and autophagic activity. PloS ONE 2014, 9, e94382. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Chen, Z.; Wang, C.; Song, L.; Zou, Y.; Zhang, L.; Hui, R.; Wang, J. Cardiac-specific overexpression of miR-222 induces heart failure and inhibits autophagy in mice. Cell. Physiol. Biochem. 2016, 39, 1503–1511. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.; Chen, J.; Wang, N.; Zhu, G.; Duan, X.; Ling, F. MiRNA-30e mediated cardioprotection of ACE2 in rats with doxorubicin-induced heart failure through inhibiting cardiomyocytes autophagy. Life Sci. 2017, 169, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Wang, A.; Xu, Y.; Qiao, S.; An, J.; Li, H.; Wang, C. Role of microRNA-1-mediated AMP-activated protein kinase pathway in cardiac fibroblasts induced by high glucose in rats. Zhonghua wei zhong bing ji jiu yi xue 2018, 30, 145–150. [Google Scholar] [PubMed]
- D’Avenia, M.; Citro, R.; De Marco, M.; Veronese, A.; Rosati, A.; Visone, R.; Leptidis, S.; Philippen, L.; Vitale, G.; Cavallo, A.; et al. A novel miR-371a-5p-mediated pathway, leading to BAG3 upregulation in cardiomyocytes in response to epinephrine, is lost in takotsubo cardiomyopathy. Cell Death Dis. 2015, 6, e1948. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Yang, L.; Wang, L.; Tang, B.; Wang, J.; Li, Q. MicroRNA-34a protects myocardial cells against ischemia-reperfusion injury through inhibiting autophagy via regulating TNAα expression. Biochem. Cell Biol. 2018, 96, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Deng, Y.; Xu, Y.; Jin, W.; Li, H. MicroRNA-223 protects neonatal rat cardiomyocytes and h9c2 cells from hypoxia-induced apoptosis and excessive autophagy via the akt/mtor pathway by targeting parp-1. J. Mol. Cell. Cardiol. 2018, 118, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Dong, Y.H.; Du, W.; Shi, C.Y.; Wang, K.; Tariq, M.A.; Wang, J.X.; Li, P.F. The role of microRNAs in myocardial infarction: From molecular mechanism to clinical application. Int. J. Mol. Sci. 2017, 18, 745. [Google Scholar] [CrossRef] [PubMed]
- Bo, L.; Su-Ling, D.; Fang, L.; Lu-Yu, Z.; Tao, A.; Stefan, D.; Kun, W.; Pei-Feng, L. Autophagic program is regulated by miR-325. Cell Death Differ. 2014, 21, 967–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Li, J.; Kou, B.; Yi, Q.; Shi, T. MicroRNA-30e protects the heart against ischemia and reperfusion injury through autophagy and the notch1/Hes1/Akt signaling pathway. Int. J. Mol. Med. 2018, 41, 3221–3230. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Xie, J.; Li, R.; Shi, J.; Sun, J.; Gu, R.; Ding, L.; Wang, L.; Xu, B. Overexpression of microRNA-99a attenuates heart remodelling and improves cardiac performance after myocardial infarction. J. Cell. Mol. Med. 2014, 18, 919–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zeng, Z.; Li, Q.; Xu, Q.; Xie, J.; Hao, H.; Luo, G.; Liao, W.; Bin, J.; Huang, X.; et al. Inhibition of microRNA-497 ameliorates anoxia/reoxygenation injury in cardiomyocytes by suppressing cell apoptosis and enhancing autophagy. Oncotarget 2015, 6, 18829–18844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Cui, T. Autophagy modulation: A potential therapeutic approach in cardiac hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 2017, 313, H304–H319. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Su, M.; Wang, S.; Zou, Y.; Wang, X.; Wang, Y.; Cui, H.; Zhao, P.; Hui, R.; Wang, J. MiR-451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting tsc1. J. Cell. Mol. Med. 2014, 18, 2266–2274. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.D.; Qin, R.H.; Yang, J.J.; Xu, S.S.; Tao, H.; Ding, X.S.; Shi, K.H. DNMT3A controls miR-200b in cardiac fibroblast autophagy and cardiac fibrosis. Inflamm. Res. 2018, 67, 681–690. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, T.; Li, M.-Y.; Li, P.-F.; Cao, J.-M. MicroRNAs in Cardiac Autophagy: Small Molecules and Big Role. Cells 2018, 7, 104. https://doi.org/10.3390/cells7080104
Sun T, Li M-Y, Li P-F, Cao J-M. MicroRNAs in Cardiac Autophagy: Small Molecules and Big Role. Cells. 2018; 7(8):104. https://doi.org/10.3390/cells7080104
Chicago/Turabian StyleSun, Teng, Meng-Yang Li, Pei-Feng Li, and Ji-Min Cao. 2018. "MicroRNAs in Cardiac Autophagy: Small Molecules and Big Role" Cells 7, no. 8: 104. https://doi.org/10.3390/cells7080104
APA StyleSun, T., Li, M. -Y., Li, P. -F., & Cao, J. -M. (2018). MicroRNAs in Cardiac Autophagy: Small Molecules and Big Role. Cells, 7(8), 104. https://doi.org/10.3390/cells7080104