When Unity Is Strength: The Strategies Used by Chlamydomonas to Survive Environmental Stresses
Abstract
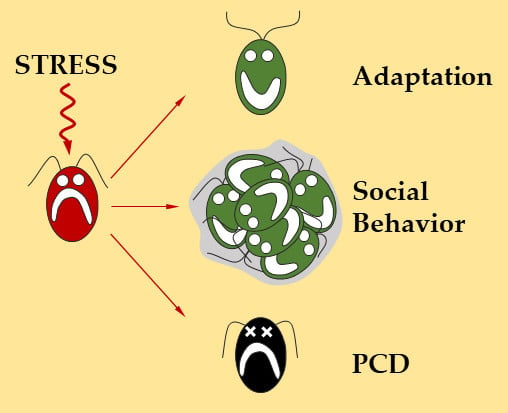
:1. Introduction
2. Coping Strategies for Moderate Stress
2.1. Acclimation: Building Defenses to Protect Cells from Future Stresses
2.2. Protection by Degradation
3. Socialization Allows for Better Resistance to Severe Stress
3.1. Multicellular Structures: Stronger Together
3.2. Programmed Cell Death: Sucide for the Good of the Community
4. Cellular Reinforcement Strategies
4.1. The Cell Wall: The Last Defender
4.2. Cyst: Surviving Extreme Conditions
4.3. Zygospore: When Sex Comes to the Rescue
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dickman, M.; Williams, B.; Li, Y.; de Figueiredo, P.; Wolpert, T. Reassessing apoptosis in plants. Nat. Plants 2017, 3, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.; Ferretti, U.; Sedlářová, M.; Pospíšil, P. Singlet oxygen production in Chlamydomonas reinhardtii under heat stress. Sci. Rep. 2016, 6, 20094. [Google Scholar] [CrossRef]
- Du, Z.-Y.; Lucker, B.F.; Zienkiewicz, K.; Miller, T.E.; Zienkiewicz, A.; Sears, B.B.; Kramer, D.M.; Benning, C. Galactoglycerolipid Lipase PGD1 Is Involved in Thylakoid Membrane Remodeling in Response to Adverse Environmental Conditions in Chlamydomonas. Plant Cell 2018, 30, 447–465. [Google Scholar] [CrossRef]
- Nagy, V.; Vidal-Meireles, A.; Podmaniczki, A.; Szentmihályi, K.; Rákhely, G.; Zsigmond, L.; Kovács, L.; Tóth, S.Z. The mechanism of photosystem-II inactivation during sulphur deprivation-induced H2 production in Chlamydomonas reinhardtii. Plant J. 2018, 94, 548–561. [Google Scholar] [CrossRef]
- Stoiber, T.L.; Shafer, M.M.; Armstrong, D.E. Induction of reactive oxygen species in Chlamydomonas reinhardtii in response to contrasting trace metal exposures. Environ. Toxicol. 2013, 28, 516–523. [Google Scholar] [CrossRef]
- Laloi, C.; Apel, K.; Danon, A. Reactive oxygen signalling: The latest news. Curr. Opin. Plant Biol. 2004, 7, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Erickson, E.; Wakao, S.; Niyogi, K.K. Light stress and photoprotection in Chlamydomonas reinhardtii. Plant J. 2015, 82, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Jokel, M.; Johnson, X.; Peltier, G.; Aro, E.-M.; Allahverdiyeva, Y. Hunting the main player enabling Chlamydomonas reinhardtii growth under fluctuating light. Plant J. 2018, 94, 822–835. [Google Scholar] [CrossRef] [PubMed]
- Ledford, H.K.; Chin, B.L.; Niyogi, K.K. Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryot. Cell 2007, 6, 919–930. [Google Scholar] [CrossRef]
- Wakao, S.; Chin, B.L.; Ledford, H.K.; Dent, R.M.; Casero, D.; Pellegrini, M.; Merchant, S.S.; Niyogi, K.K. Phosphoprotein SAK1 is a regulator of acclimation to singlet oxygen in Chlamydomonas reinhardtii. eLife 2014, 3, e02286. [Google Scholar] [CrossRef]
- Fischer, B.B.; Eggen, R.I.; Niyogi, K.K. Characterization of singlet oxygen-accumulating mutants isolated in a screen for altered oxidative stress response in Chlamydomonas reinhardtii. BMC Plant. Biol. 2010, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Tilbrook, K.; Dubois, M.; Crocco, C.D.; Yin, R.; Chappuis, R.; Allorent, G.; Schmid-Siegert, E.; Goldschmidt-Clermont, M.; Ulm, R. UV-B Perception and Acclimation in Chlamydomonas reinhardtii. Plant Cell 2016, 28, 966–983. [Google Scholar] [CrossRef] [PubMed]
- Meijer, H.J.G.; van Himbergen, J.A.J.; Musgrave, A.; Munnik, T. Acclimation to salt modifies the activation of several osmotic stress-activated lipid signalling pathways in Chlamydomonas. Phytochemistry 2017, 135, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Charles, E.D.; Muhamadali, H.; Goodacre, R.; Pittman, J.K. Biochemical signatures of acclimation by Chlamydomonas reinhardtii to different ionic stresses. Algal Res. 2019, 37, 83–91. [Google Scholar] [CrossRef]
- Laloi, C.; Stachowiak, M.; Pers-Kamczyc, E.; Warzych, E.; Murgia, I.; Apel, K. Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 672–677. [Google Scholar] [CrossRef]
- Fischer, B.B.; Ledford, H.K.; Wakao, S.; Huang, S.G.; Casero, D.; Pellegrini, M.; Merchant, S.S.; Koller, A.; Eggen, R.I.L.; Niyogi, K.K. SINGLET OXYGEN RESISTANT 1 links reactive electrophile signaling to singlet oxygen acclimation in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2012, 109, E1302–E1311. [Google Scholar] [CrossRef]
- Liang, T.; Yang, Y.; Liu, H. Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytol. 2019, 221, 1247–1252. [Google Scholar] [CrossRef]
- Budenholzer, L.; Cheng, C.L.; Li, Y.; Hochstrasser, M. Proteasome Structure and Assembly. J. Mol. Biol. 2017, 429, 3500–3524. [Google Scholar] [CrossRef]
- Smalle, J.; Vierstra, R.D. The Ubiquitin 26s Proteasome Proteolytic Pathway. Annu. Rev. Plant. Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef]
- Heredia-Martínez, L.G.; Andrés-Garrido, A.; Martínez-Force, E.; Pérez-Pérez, M.E.; Crespo, J.L. Chloroplast Damage Induced by the Inhibition of Fatty Acid Synthesis Triggers Autophagy in Chlamydomonas. Plant. Physiol. 2018, 178, 1112–1129. [Google Scholar] [CrossRef]
- Valledor, L.; Furuhashi, T.; Hanak, A.-M.; Weckwerth, W. Systemic Cold Stress Adaptation of Chlamydomonas reinhardtii. Mol. Cell. Proteomics 2013, 12, 2032–2047. [Google Scholar] [CrossRef] [PubMed]
- Jamers, A.; Van der Ven, K.; Moens, L.; Robbens, J.; Potters, G.; Guisez, Y.; Blust, R.; De Coen, W. Effect of copper exposure on gene expression profiles in Chlamydomonas reinhardtii based on microarray analysis. Aquat. Toxicol. 2006, 80, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Chamari, W.; Harry, K.; Belinda, W.; Robert, W. Differential Proteome Analysis of Chlamydomonas reinhardtii Response to Arsenic Exposure. Am. J. Plant. Sci. 2012, 3, 764. [Google Scholar]
- Simon, D.F.; Domingos, R.F.; Hauser, C.; Hutchins, C.M.; Zerges, W.; Wilkinson, K.J. Transcriptome sequencing (RNA-seq) analysis of the effects of metal nanoparticle exposure on the transcriptome of Chlamydomonas reinhardtii. Appl. Environ. Microbiol. 2013, 79, 4774–4785. [Google Scholar] [CrossRef] [PubMed]
- Vallentine, P.; Hung, C.-Y.; Xie, J.; Van Hoewyk, D. The ubiquitin–proteasome pathway protects Chlamydomonas reinhardtii against selenite toxicity, but is impaired as reactive oxygen species accumulate. AoB Plants 2014, 6, plu062. [Google Scholar] [CrossRef] [PubMed]
- Shang, F.; Taylor, A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic. Biol. Med. 2011, 51, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L. Role of the Ubiquitin Proteasome System in Plant Response to Abiotic Stress. In International Review of Cell and Molecular Biology; Galluzzi, L., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 343, pp. 65–110. [Google Scholar]
- Lyzenga, W.J.; Stone, S.L. Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 2012, 63, 599–616. [Google Scholar] [CrossRef]
- Wang, C.; Youle, R.J. The Role of Mitochondria in Apoptosis. Annu. Rev. Genet. 2009, 43, 95–118. [Google Scholar] [CrossRef] [Green Version]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef]
- Avin-Wittenberg, T. Autophagy and its role in plant abiotic stress management. Plant. Cell Environ. 2019, 42, 1045–1053. [Google Scholar] [CrossRef]
- Izumi, M.; Ishida, H.; Nakamura, S.; Hidema, J. Entire Photodamaged Chloroplasts Are Transported to the Central Vacuole by Autophagy. Plant. Cell 2017, 29, 377–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broda, M.; Millar, A.H.; Van Aken, O. Mitophagy: A Mechanism for Plant Growth and Survival. Trends Plant. Sci. 2018, 23, 434–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kajikawa, M.; Yamauchi, M.; Shinkawa, H.; Tanaka, M.; Hatano, K.; Nishimura, Y.; Kato, M.; Fukuzawa, H. Isolation and Characterization of Chlamydomonas Autophagy-Related Mutants in Nutrient-Deficient Conditions. Plant. Cell Physiol. 2019, 60, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, M.E.; Couso, I.; Heredia-Martínez, L.G.; Crespo, J.L. Monitoring Autophagy in the Model Green Microalga Chlamydomonas reinhardtii. Cells 2017, 6, 36. [Google Scholar] [CrossRef]
- Wesselborg, S.; Stork, B. Autophagy signal transduction by ATG proteins: From hierarchies to networks. Cell. Mol. Life Sci. 2015, 72, 4721–4757. [Google Scholar] [CrossRef]
- Pérez-Pérez, M.E.; Florencio, F.J.; Crespo, J.L. Inhibition of Target of Rapamycin Signaling and Stress Activate Autophagy in Chlamydomonas reinhardtii. Plant. Physiol. 2010, 152, 1874–1888. [Google Scholar] [CrossRef]
- Goodenough, U.; Blaby, I.; Casero, D.; Gallaher, S.D.; Goodson, C.; Johnson, S.; Lee, J.-H.; Merchant, S.S.; Pellegrini, M.; Roth, R.; et al. The Path to Triacylglyceride Obesity in the sta6 Strain of Chlamydomonas reinhardtii. Eukaryot. Cell 2014, 13, 591–613. [Google Scholar] [CrossRef]
- Pérez-Martín, M.; Pérez-Pérez, M.E.; Lemaire, S.D.; Crespo, J.L. Oxidative Stress Contributes to Autophagy Induction in Response to Endoplasmic Reticulum Stress in Chlamydomonas reinhardtii. Plant. Physiol. 2014, 166, 997–1008. [Google Scholar] [CrossRef]
- Tran, Q.-G.; Cho, K.; Park, S.-B.; Kim, U.; Lee, Y.J.; Kim, H.-S. Impairment of starch biosynthesis results in elevated oxidative stress and autophagy activity in Chlamydomonas reinhardtii. Sci. Rep. 2019, 9, 9856. [Google Scholar] [CrossRef]
- Pérez-Martín, M.; Blaby-Haas, C.E.; Pérez-Pérez, M.E.; Andrés-Garrido, A.; Blaby, I.K.; Merchant, S.S.; Crespo, J.L. Activation of Autophagy by Metals in Chlamydomonas reinhardtii. Eukaryot. Cell 2015, 14, 964–973. [Google Scholar] [CrossRef]
- Pérez-Pérez, M.E.; Couso, I.; Crespo, J.L. Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy 2012, 8, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, M.E.; Lemaire, S.D.; Crespo, J.L. Reactive Oxygen Species and Autophagy in Plants and Algae. Plant. Physiol. 2012, 160, 156–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Pérez, M.E.; Zaffagnini, M.; Marchand, C.H.; Crespo, J.L.; Lemaire, S.D. The yeast autophagy protease Atg4 is regulated by thioredoxin. Autophagy 2014, 10, 1953–1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Pérez, M.E.; Lemaire, S.D.; Crespo, J.L. Control of Autophagy in Chlamydomonas Is Mediated through Redox-Dependent Inactivation of the ATG4 Protease. Plant. Physiol. 2016, 172, 2219–2234. [Google Scholar] [CrossRef] [PubMed]
- Harris, E.H. The Chlamydomonas Sourcebook; Academic Press: San Diego, CA, USA, 1989; ISBN 978-0-12-326880-8. [Google Scholar]
- Iwasa, K.; Murakami, S. Palmelloid Formation of Chlamydomonas I. Palmelloid Induction by Organic Acids. Physiol. Plant. 1968, 21, 1224–1233. [Google Scholar] [CrossRef]
- Pančić, M.; Kiørboe, T. Phytoplankton defence mechanisms: Traits and trade-offs. Biol. Rev. 2018, 93, 1269–1303. [Google Scholar] [CrossRef]
- Fisher, R.M.; Bell, T.; West, S.A. Multicellular group formation in response to predators in the alga Chlorella vulgaris. J. Evol. Biol. 2016, 29, 551–559. [Google Scholar] [CrossRef]
- Lurling, M.; Beekman, W. Palmelloids formation in Chlamydomonas reinhardtii: Defence against rotifer predators? Ann. Limnol. Int. J. Limnol. 2006, 42, 65–72. [Google Scholar] [CrossRef]
- Iwasa, K.; Murakami, S. Palmelloid Formation of Chlamydomonas II. Mechanism of Palmelloid Formation by Organic Acids. Physiol. Plant. 1969, 22, 43–50. [Google Scholar] [CrossRef]
- Olsen, Y.; Knutsen, G.; Lien, T. Characteristics of Phosphorus Limitation in Chlamydomonas reinhardtii (chlorophyceae) and Its Palmelloids. J. Phycol. 1983, 19, 313–319. [Google Scholar] [CrossRef]
- Samadani, M.; Dewez, D. Cadmium accumulation and toxicity affect the extracytoplasmic polyphosphate level in Chlamydomonas reinhardtii. Ecotoxicol. Environ. Saf. 2018, 166, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Takouridis, S.J.; Tribe, D.E.; Gras, S.L.; Martin, G.J.O. The selective breeding of the freshwater microalga Chlamydomonas reinhardtii for growth in salinity. Bioresour. Technol. 2015, 184, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Khona, D.K.; Shirolikar, S.M.; Gawde, K.K.; Hom, E.; Deodhar, M.A.; D’Souza, J.S. Characterization of salt stress-induced palmelloids in the green alga, Chlamydomonas reinhardtii. Algal Res. 2016, 16, 434–448. [Google Scholar] [CrossRef]
- Neelam, S.; Subramanyam, R. Alteration of photochemistry and protein degradation of photosystem II from Chlamydomonas reinhardtii under high salt grown cells. J. Photochem. Photobiol. B 2013, 124, 63–70. [Google Scholar] [CrossRef]
- Visviki, I.; Santikul, D. The pH tolerance of Chlamydomonas applanata (Volvocales, Chlorophyta). Arch. Environ. Contam. Toxicol. 2000, 38, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Bray, D.F.; Wagenaar, E.B. Ultrastructure of Chlamydomonas eugametos palmelloids induced by chloroplatinic acid treatment. J. Bacteriol. 1975, 121, 338–343. [Google Scholar]
- Nakamura, K.; Sakon, M.; Hatanaka, M.K. Chemical Factors Affecting Palmelloid-Forming Activity of Chloroplatinic Acid on Chlamydomonas eugametos. Physiol. Plant. 1976, 36, 293–296. [Google Scholar] [CrossRef]
- Sathe, S.; Durand, P.M. Cellular aggregation in Chlamydomonas (Chlorophyceae) is chimaeric and depends on traits like cell size and motility. Eur. J. Phycol. 2016, 51, 129–138. [Google Scholar] [CrossRef]
- Fan, J.; Zheng, L.; Bai, Y.; Saroussi, S.; Grossman, A.R. Flocculation of Chlamydomonas reinhardtii with Different Phenotypic Traits by Metal Cations and High pH. Front. Plant. Sci. 2017, 8, 1997. [Google Scholar] [CrossRef]
- Goff, K.L.; Headley, J.V.; Lawrence, J.R.; Wilson, K.E. Assessment of the effects of oil sands naphthenic acids on the growth and morphology of Chlamydomonas reinhardtii using microscopic and spectromicroscopic techniques. Sci. Total Environ. 2013, 442, 116–122. [Google Scholar] [CrossRef]
- Day, J.G.; Gong, Y.; Hu, Q. Microzooplanktonic grazers—A potentially devastating threat to the commercial success of microalgal mass culture. Algal Res. 2017, 27, 356–365. [Google Scholar] [CrossRef]
- Porter, K.G. Viable gut passage of gelatinous green algae ingested by Daphnia. SIL Proc. 1922–2010 1975, 19, 2840–2850. [Google Scholar] [CrossRef]
- Wilson, W.W.; Wade, M.M.; Holman, S.C.; Champlin, F.R. Status of methods for assessing bacterial cell surface charge properties based on zeta potential measurements. J. Microbiol. Methods 2001, 43, 153–164. [Google Scholar] [CrossRef]
- Banerjee, C.; Ghosh, S.; Sen, G.; Mishra, S.; Shukla, P.; Bandopadhyay, R. Study of algal biomass harvesting through cationic cassia gum, a natural plant based biopolymer. Bioresour. Technol. 2014, 151, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Santos, E.; Vila, M.; de la Vega, M.; León, R.; Vigara, J. Study of bioflocculation induced by Saccharomyces bayanus var. uvarum and flocculating protein factors in microalgae. Algal Res. 2015, 8, 23–29. [Google Scholar]
- Ogata, T.; Izumikawa, M.; Kohno, K.; Shibata, K. Chromosomal location of Lg-FLO1 in bottom-fermenting yeast and the FLO5 locus of industrial yeast. J. Appl. Microbiol. 2008, 105, 1186–1198. [Google Scholar] [CrossRef] [PubMed]
- Soares, E.V. Flocculation in Saccharomyces cerevisiae: A review. J. Appl. Microbiol. 2011, 110, 1–18. [Google Scholar] [CrossRef]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef]
- Díaz-Santos, E.; Vila, M.; Vigara, J.; León, R. A new approach to express transgenes in microalgae and its use to increase the flocculation ability of Chlamydomonas reinhardtii. J. Appl. Phycol. 2016, 28, 1611–1621. [Google Scholar] [CrossRef]
- Musgrave, A.; van der Steuyt, P.; Ero, L. Concanavalin A binding to Chlamydomonas eugametos flagellar proteins and its effect on sexual reproduction. Planta 1979, 147, 51–56. [Google Scholar] [CrossRef]
- Millikin, B.E.; Weiss, R.L. Localization of concanavalin A binding carbohydrate in Chlamydomonas flagella. J. Cell Sci. 1984, 68, 211–226. [Google Scholar] [PubMed]
- Boyd, M.; Rosenzweig, F.; Herron, M.D. Analysis of motility in multicellular Chlamydomonas reinhardtii evolved under predation. PLoS ONE 2018, 13, e0192184. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, G.L.; Miranda-Saavedra, D.; Barton, G.J. Genome Analysis of the Unicellular Green Alga Chlamydomonas reinhardtii Indicates an Ancient Evolutionary Origin for Key Pattern Recognition and Cell-Signaling Protein Families. Genetics 2008, 179, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Lowder, L.G.; Herbert, S.K. Heterologous expression of a Volvox cell adhesion molecule causes flocculation in Chlamydomonas reinhardtii. J. Appl. Phycol. 2015, 27, 721–731. [Google Scholar] [CrossRef]
- Seifert, G.J. Fascinating Fasciclins: A Surprisingly Widespread Family of Proteins that Mediate Interactions between the Cell Exterior and the Cell Surface. Int. J. Mol. Sci. 2018, 19, 1628. [Google Scholar] [CrossRef]
- Johnson, K.L.; Jones, B.J.; Bacic, A.; Schultz, C.J. The Fasciclin-Like Arabinogalactan Proteins of Arabidopsis. A Multigene Family of Putative Cell Adhesion Molecules. Plant. Physiol. 2003, 133, 1911–1925. [Google Scholar] [CrossRef] [Green Version]
- Grosberg, R.K.; Strathmann, R.R. The Evolution of Multicellularity: A Minor Major Transition? Annu. Rev. Ecol. Evol. Syst. 2007, 38, 621–654. [Google Scholar] [CrossRef] [Green Version]
- Prochnik, S.E.; Umen, J.; Nedelcu, A.M.; Hallmann, A.; Miller, S.M.; Nishii, I.; Ferris, P.; Kuo, A.; Mitros, T.; Fritz-Laylin, L.K.; et al. Genomic Analysis of Organismal Complexity in the Multicellular Green Alga Volvox carteri. Science 2010, 329, 223–226. [Google Scholar] [CrossRef]
- Olson, B.J. From brief encounters to lifelong unions. eLife 2013, 2, e01893. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Herron, M.D.; Howell, K.; Pentz, J.T.; Rosenzweig, F.; Travisano, M. Experimental evolution of an alternating uni- and multicellular life cycle in Chlamydomonas reinhardtii. Nat. Commun. 2013, 4, 2742. [Google Scholar] [CrossRef]
- Herron, M.D.; Borin, J.M.; Boswell, J.C.; Walker, J.; Chen, I.-C.K.; Knox, C.A.; Boyd, M.; Rosenzweig, F.; Ratcliff, W.C. De novo origins of multicellularity in response to predation. Sci. Rep. 2019, 9, 2328. [Google Scholar] [CrossRef]
- Boraas, M.E.; Seale, D.B.; Boxhorn, J.E. Phagotrophy by a flagellate selects for colonial prey: A possible origin of multicellularity. Evol. Ecol. 1998, 12, 153–164. [Google Scholar] [CrossRef]
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef]
- Earnshaw, W.C.; Martins, L.M.; Kaufmann, S.H. Mammalian Caspases: Structure, Activation, Substrates, and Functions During Apoptosis. Annu. Rev. Biochem. 1999, 68, 383–424. [Google Scholar] [CrossRef]
- He, B.; Lu, N.; Zhou, Z. Cellular and Nuclear Degradation during Apoptosis. Curr. Opin. Cell Biol. 2009, 21, 900–912. [Google Scholar] [CrossRef]
- Zhivotosky, B.; Orrenius, S. Assessment of apoptosis and necrosis by DNA fragmentation and morphological criteria. Curr. Protoc. Cell Biol. 2001, 12, 18.3.1–18.3.23. [Google Scholar]
- Fuchs, Y.; Steller, H. Programmed Cell Death in Animal Development and Disease. Cell 2011, 147, 742–758. [Google Scholar] [CrossRef] [Green Version]
- Bayles, K.W. Bacterial programmed cell death: Making sense of a paradox. Nat. Rev. Microbiol. 2014, 12, 63–69. [Google Scholar] [CrossRef]
- Bidle, K.D. Programmed Cell Death in Unicellular Phytoplankton. Curr. Biol. 2016, 26, R594–R607. [Google Scholar] [CrossRef] [Green Version]
- Carmona-Gutierrez, D.; Eisenberg, T.; Büttner, S.; Meisinger, C.; Kroemer, G.; Madeo, F. Apoptosis in yeast: Triggers, pathways, subroutines. Cell Death Differ. 2010, 17, 763–773. [Google Scholar] [CrossRef]
- Sirisha, V.L.; Sinha, M.; D’Souza, J.S. Menadione-induced caspase-dependent programmed cell death in the green chlorophyte Chlamydomonas reinhardtii. J. Phycol. 2014, 50, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Yordanova, Z.P.; Woltering, E.J.; Kapchina-Toteva, V.M.; Iakimova, E.T. Mastoparan-induced programmed cell death in the unicellular alga Chlamydomonas reinhardtii. Ann. Bot. 2013, 111, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Vavilala, S.L.; Gawde, K.K.; Sinha, M.; D’Souza, J.S. Programmed cell death is induced by hydrogen peroxide but not by excessive ionic stress of sodium chloride in the unicellular green alga Chlamydomonas reinhardtii. Eur. J. Phycol. 2015, 50, 422–438. [Google Scholar] [CrossRef]
- Ge, Y.; Cai, Y.-M.; Bonneau, L.; Rotari, V.; Danon, A.; McKenzie, E.A.; McLellan, H.; Mach, L.; Gallois, P. Inhibition of cathepsin B by caspase-3 inhibitors blocks programmed cell death in Arabidopsis. Cell Death Differ. 2016, 23, 1493–1501. [Google Scholar] [CrossRef]
- Hatsugai, N.; Yamada, K.; Goto-Yamada, S.; Hara-Nishimura, I. Vacuolar processing enzyme in plant programmed cell death. Front. Plant. Sci. 2015, 6, 234. [Google Scholar] [CrossRef]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; Van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis type I metacaspases control cell death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef]
- Tsiatsiani, L.; Van Breusegem, F.; Gallois, P.; Zavialov, A.; Lam, E.; Bozhkov, P.V. Metacaspases. Cell Death Differ. 2011, 18, 1279–1288. [Google Scholar] [CrossRef]
- Murik, O.; Elboher, A.; Kaplan, A. Dehydroascorbate: A possible surveillance molecule of oxidative stress and programmed cell death in the green alga Chlamydomonas reinhardtii. New Phytol. 2014, 202, 471–484. [Google Scholar] [CrossRef]
- Zuo, Z.; Zhu, Y.; Bai, Y.; Wang, Y. Acetic acid-induced programmed cell death and release of volatile organic compounds in Chlamydomonas reinhardtii. Plant. Physiol. Biochem. PPB 2012, 51, 175–184. [Google Scholar] [CrossRef]
- Moharikar, S.; D’Souza, J.S.; Kulkarni, A.B.; Rao, B.J. Apoptotic-Like Cell Death Pathway Is Induced in Unicellular Chlorophyte Chlamydomonas reinhardtii (chlorophyceae) Cells Following Uv Irradiation: Detection and Functional Analyses1. J. Phycol. 2006, 42, 423–433. [Google Scholar] [CrossRef]
- Durand, P.M.; Rashidi, A.; Michod, R.E. How an organism dies affects the fitness of its neighbors. Am. Nat. 2011, 177, 224–232. [Google Scholar] [CrossRef]
- Ferris, P.J.; Woessner, J.P.; Waffenschmidt, S.; Kilz, S.; Drees, J.; Goodenough, U.W. Glycosylated Polyproline II Rods with Kinks as a Structural Motif in Plant Hydroxyproline-Rich Glycoproteins. Biochemistry 2001, 40, 2978–2987. [Google Scholar] [CrossRef]
- Macfie, S.M.; Tarmohamed, Y.; Welbourn, P.M. Effects of cadmium, cobalt, copper, and nickel on growth of the green alga Chlamydomonas reinhardtii: The influences of the cell wall and pH. Arch. Environ. Contam. Toxicol. 1994, 27, 454–458. [Google Scholar] [CrossRef]
- Macfie, S.M.; Welbourn, P.M. The Cell Wall as a Barrier to Uptake of Metal Ions in the Unicellular Green Alga Chlamydomonas reinhardtii (Chlorophyceae). Arch. Environ. Contam. Toxicol. 2000, 39, 413–419. [Google Scholar] [CrossRef]
- Prasad, M.N.V.; Drej, K.; Skawińska, A.; Strałka, K. Toxicity of Cadmium and Copper in Chlamydomonas reinhardtii Wild-Type (WT 2137) and Cell Wall Deficient Mutant Strain (CW 15). Bull. Environ. Contam. Toxicol. 1998, 60, 306–311. [Google Scholar] [CrossRef]
- Baba, M.; Suzuki, I.; Shiraiwa, Y. Proteomic analysis of high-CO(2)-inducible extracellular proteins in the unicellular green alga, Chlamydomonas reinhardtii. Plant. Cell Physiol. 2011, 52, 1302–1314. [Google Scholar] [CrossRef]
- Takahashi, H.; Braby, C.E.; Grossman, A.R. Sulfur Economy and Cell Wall Biosynthesis during Sulfur Limitation of Chlamydomonas reinhardtii. Plant. Physiol. 2001, 127, 665–673. [Google Scholar] [CrossRef]
- Davies, J.P.; Yildiz, F.H.; Grossman, A. Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J. 1996, 15, 2150–2159. [Google Scholar] [CrossRef]
- Van Donk, E.; LÜrling, M.; Hessen, D.O.; Lokhorst, G.M. Altered cell wall morphology in nutrient-deficient phytoplankton and its impact on grazers. Limnol. Oceanogr. 1997, 42, 357–364. [Google Scholar] [CrossRef] [Green Version]
- van Donk, E.; Hessen, D.O. Grazing resistance in nutrient-stressed phytoplankton. Oecologia 1993, 93, 508–511. [Google Scholar] [CrossRef]
- Lürling, M.; Van Donk, E. Life history consequences for Daphnia pulex feeding on nutrient-limited phytoplankton. Freshw. Biol. 1997, 38, 693–709. [Google Scholar] [CrossRef]
- Porter, K.G. Selective Grazing and Differential Digestion of Algae by Zooplankton. Nature 1973, 244, 179. [Google Scholar] [CrossRef]
- Scholz, M.; Hoshino, T.; Johnson, D.; Riley, M.R.; Cuello, J. Flocculation of wall-deficient cells of Chlamydomonas reinhardtii mutant cw15 by calcium and methanol. Biomass Bioenergy 2011, 35, 4835–4840. [Google Scholar] [CrossRef]
- Mini, P.; Demurtas, O.C.; Valentini, S.; Pallara, P.; Aprea, G.; Ferrante, P.; Giuliano, G. Agrobacterium-mediated and electroporation-mediated transformation of Chlamydomonas reinhardtii: A comparative study. BMC Biotechnol. 2018, 18, 11. [Google Scholar] [CrossRef]
- Segawa, T.; Matsuzaki, R.; Takeuchi, N.; Akiyoshi, A.; Navarro, F.; Sugiyama, S.; Yonezawa, T.; Mori, H. Bipolar dispersal of red-snow algae. Nat. Commun. 2018, 9, 3094. [Google Scholar] [CrossRef]
- Darwin, C. Journal of Researches into the Natural History and Geology of the Countries Visited during the Voyage of HMS Beagle Round the World, under the Command of Capt. Fitz Roy, R.N., 2nd ed.; Cambridge University Press: London, UK, 1860. [Google Scholar]
- Procházková, L.; Remias, D.; Holzinger, A.; Řezanka, T.; Nedbalová, L. Ecophysiological and morphological comparison of two populations of Chlainomonas sp. (Chlorophyta) causing red snow on ice-covered lakes in the High Tatras and Austrian Alps. Eur. J. Phycol. 2018, 53, 230–243. [Google Scholar] [CrossRef]
- Müller, T.; Bleiß, W.; Martin, C.-D.; Rogaschewski, S.; Fuhr, G. Snow algae from northwest Svalbard: Their identification, distribution, pigment and nutrient content. Polar Biol. 1998, 20, 14–32. [Google Scholar] [CrossRef]
- Remias, D.; Lütz-Meindl, U.; Lütz, C. Photosynthesis, pigments and ultrastructure of the alpine snow alga Chlamydomonas nivalis. Eur. J. Phycol. 2005, 40, 259–268. [Google Scholar] [CrossRef]
- Gorton, H.L.; Vogelmann, T.C. Ultraviolet Radiation and the Snow Alga Chlamydomonas nivalis (Bauer) Wille. Photochem. Photobiol. 2003, 77, 608–615. [Google Scholar] [CrossRef]
- Boussiba, S. Carotenogenesis in the green alga Haematococcus pluvialis: Cellular physiology and stress response. Physiol. Plant. 2000, 108, 111–117. [Google Scholar] [CrossRef]
- Kakizono, T.; Kobayashi, M.; Nagai, S. Effect of carbon/nitrogen ratio on encystment accompanied with astaxanthin formation in a green alga, Haematococcus pluvialis. J. Ferment. Bioeng. 1992, 74, 403–405. [Google Scholar] [CrossRef]
- Li, Y.; Sommerfeld, M.; Chen, F.; Hu, Q. Effect of photon flux densities on regulation of carotenogenesis and cell viability of Haematococcus pluvialis (Chlorophyceae). J. Appl. Phycol. 2010, 22, 253–263. [Google Scholar] [CrossRef]
- Li, Y.; Sommerfeld, M.; Chen, F.; Hu, Q. Consumption of oxygen by astaxanthin biosynthesis: A protective mechanism against oxidative stress in Haematococcus pluvialis (Chlorophyceae). J. Plant. Physiol. 2008, 165, 1783–1797. [Google Scholar] [CrossRef]
- Sarada, R.; Tripathi, U.; Ravishankar, G.A. Influence of stress on astaxanthin production in Haematococcus pluvialis grown under different culture conditions. Process. Biochem. 2002, 37, 623–627. [Google Scholar] [CrossRef]
- Tjahjono, A.E.; Hayama, Y.; Kakizono, T.; Terada, Y.; Nishio, N.; Nagai, S. Hyper-accumulation of astaxanthin in a green alga Haematococcus pluvialis at elevated temperatures. Biotechnol. Lett. 1994, 16, 133–138. [Google Scholar] [CrossRef]
- Rezanka, T.; Nedbalová, L.; Sigler, K.; Cepák, V. Identification of astaxanthin diglucoside diesters from snow alga Chlamydomonas nivalis by liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. Phytochemistry 2008, 69, 479–490. [Google Scholar] [CrossRef]
- Grossman, A.R.; Lohr, M.; Im, C.S. Chlamydomonas reinhardtii in the Landscape of Pigments. Annu. Rev. Genet. 2004, 38, 119–173. [Google Scholar] [CrossRef]
- Lemoine, Y.; Schoefs, B. Secondary ketocarotenoid astaxanthin biosynthesis in algae: A multifunctional response to stress. Photosynth. Res. 2010, 106, 155–177. [Google Scholar] [CrossRef]
- Bidigare, R.R.; Ondrusek, M.E.; Kennicutt, M.C.; Iturriaga, R.; Harvey, H.R.; Hoham, R.W.; Macko, S.A. Evidence for a photoprotective function for secondary carotenoids of snow algae. J. Phycol. 1993, 29, 427–434. [Google Scholar] [CrossRef]
- Scibilia, L.; Girolomoni, L.; Berteotti, S.; Alboresi, A.; Ballottari, M. Photosynthetic response to nitrogen starvation and high light in Haematococcus pluvialis. Algal Res. 2015, 12, 170–181. [Google Scholar] [CrossRef]
- Perozeni, F.; Cazzaniga, S.; Baier, T.; Zanoni, F.; Zoccatelli, G.; Lauersen, K.J.; Wobbe, L.; Ballottari, M. Turning a green alga red: Engineering astaxanthin biosynthesis by intragenic pseudogene revival in Chlamydomonas reinhardtii. bioRxiv 2019, 535989. [Google Scholar] [CrossRef]
- Strenkert, D.; Schmollinger, S.; Gallaher, S.D.; Salomé, P.A.; Purvine, S.O.; Nicora, C.D.; Mettler-Altmann, T.; Soubeyrand, E.; Weber, A.P.M.; Lipton, M.S.; et al. Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proc. Natl. Acad. Sci. USA 2019, 116, 2374–2383. [Google Scholar] [CrossRef]
- Zou, Y.; Wenzel, S.; Müller, N.; Prager, K.; Jung, E.-M.; Kothe, E.; Kottke, T.; Mittag, M. An Animal-Like Cryptochrome Controls the Chlamydomonas Sexual Cycle. Plant. Physiol. 2017, 174, 1334–1347. [Google Scholar] [CrossRef]
- Nedelcu, A.M.; Marcu, O.; Michod, R.E. Sex as a response to oxidative stress: A twofold increase in cellular reactive oxygen species activates sex genes. Proc. R. Soc. B Biol. Sci. 2004, 271, 1591–1596. [Google Scholar] [CrossRef]
- Waffenschmidt, S.; Woessner, J.P.; Beer, K.; Goodenough, U.W. Isodityrosine cross-linking mediates insolubilization of cell walls in Chlamydomonas. Plant. Cell 1993, 5, 809–820. [Google Scholar]
- Waffenschmidt, S.; Kusch, T.; Woessner, J.P. A Transglutaminase Immunologically Related to Tissue Transglutaminase Catalyzes Cross-Linking of Cell Wall Proteins in Chlamydomonas reinhardtii. Plant. Physiol. 1999, 121, 1003–1015. [Google Scholar] [CrossRef]
- Lopez, D.; Hamaji, T.; Kropat, J.; De Hoff, P.; Morselli, M.; Rubbi, L.; Fitz-Gibbon, S.; Gallaher, S.D.; Merchant, S.S.; Umen, J.; et al. Dynamic Changes in the Transcriptome and Methylome of Chlamydomonas reinhardtii throughout Its Life Cycle. Plant. Physiol. 2015, 169, 2730–2743. [Google Scholar] [CrossRef]
- Joo, S.; Nishimura, Y.; Cronmiller, E.; Hong, R.H.; Kariyawasam, T.; Wang, M.H.; Shao, N.C.; El Akkad, S.-E.-D.; Suzuki, T.; Higashiyama, T.; et al. Gene Regulatory Networks for the Haploid-to-Diploid Transition of Chlamydomonas reinhardtii. Plant. Physiol. 2017, 175, 314–332. [Google Scholar] [CrossRef]
- Suzuki, L.; Woessner, J.P.; Uchida, H.; Kuroiwa, H.; Yuasa, Y.; Waffenschmidt, S.; Goodenough, U.W.; Kuroiwa, T. A Zygote-Specific Protein with Hydroxyproline-Rich Glycoprotein Domains and Lectin-Like Domains Involved in the Assembly of the Cell Wall of Chlamydomonas reinhardtii (chlorophyta). J. Phycol. 2000, 36, 571–583. [Google Scholar] [CrossRef]
- Heimerl, N.; Hommel, E.; Westermann, M.; Meichsner, D.; Lohr, M.; Hertweck, C.; Grossman, A.R.; Mittag, M.; Sasso, S. A giant type I polyketide synthase participates in zygospore maturation in Chlamydomonas reinhardtii. Plant. J. 2018, 95, 268–281. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Electron microscopy of zygospore formation in Chlamydomonas reinhardii. Protoplasma 1976, 87, 297–315. [Google Scholar] [CrossRef]
- VanWinkle-Swift, K.P.; Rickoll, W.L. The Zygospore Wall of Chlamydomonas Monoica (chlorophyceae): Morphogenesis and Evidence for the Presence of Sporopollenin1. J. Phycol. 1997, 33, 655–665. [Google Scholar] [CrossRef]
- Suzuki, L.; Johnson, C. Photoperiodic control of germination in the unicell Chlamydomonas. Naturwissenschaften 2002, 89, 214–220. [Google Scholar] [CrossRef]
- Trainor, F.R.; Gladych, R. Survival of algae in a desiccated soil: A 35-year study. Phycologia 1995, 34, 191–192. [Google Scholar] [CrossRef]
- Suzuki, L.; Yuasa, Y.; Kuroiwa, T. Transcription and Translation involved in Pellicle Formation in the Chlamydomonas reinhardtii Zygote. Cytologia 1997, 62, 421–425. [Google Scholar] [CrossRef]
- Avnimelech, Y.; Troeger, B.W.; Reed, L.W. Mutual Flocculation of Algae and Clay: Evidence and Implications. Science 1982, 216, 63–65. [Google Scholar] [CrossRef]
- Salim, S.; Bosma, R.; Vermuë, M.H.; Wijffels, R.H. Harvesting of microalgae by bio-flocculation. J. Appl. Phycol. 2011, 23, 849–855. [Google Scholar] [CrossRef]


| Behavior | Stress | Conditions | Reference |
|---|---|---|---|
| Palmelloids | Predator | Brachionus calyciflorus | [50] |
| Organic acids (succinate, fumarate, aspartate, glutamate, glycolate, citrate, phthalate) | 0.15–5% | [47] | |
| EDTA, GEDTA | 1.25 mM | [51] | |
| Calcium deficiency | <3.5 µM | ||
| Phosphorous deficiency | <1 µg/L | [52] | |
| Cadmium | 200–400 µM | [53] | |
| NaCl | 300–700 mM | [54] | |
| 100–150 mM | [55] | ||
| 50–150 mM | [56] | ||
| Acidic pH | pH 4.4 | [57] | |
| Chloroplatinic acid | 50 µM | [58,59] | |
| Aggregates | Predator | Brachionus calyciflorus | [50] |
| Peranema trichophorum | [60] | ||
| Acidic pH | pH 3.4 | [57] | |
| pH 2.5–pH 4 | [61] | ||
| Basic pH | pH 10–pH 13 | ||
| FeCl3, CaCl2, MgCl2 | 1–10 mM | ||
| Naphthenic acids | 100 mg/L | [62] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Carpentier, F.; Lemaire, S.D.; Danon, A. When Unity Is Strength: The Strategies Used by Chlamydomonas to Survive Environmental Stresses. Cells 2019, 8, 1307. https://doi.org/10.3390/cells8111307
de Carpentier F, Lemaire SD, Danon A. When Unity Is Strength: The Strategies Used by Chlamydomonas to Survive Environmental Stresses. Cells. 2019; 8(11):1307. https://doi.org/10.3390/cells8111307
Chicago/Turabian Stylede Carpentier, Félix, Stéphane D. Lemaire, and Antoine Danon. 2019. "When Unity Is Strength: The Strategies Used by Chlamydomonas to Survive Environmental Stresses" Cells 8, no. 11: 1307. https://doi.org/10.3390/cells8111307
APA Stylede Carpentier, F., Lemaire, S. D., & Danon, A. (2019). When Unity Is Strength: The Strategies Used by Chlamydomonas to Survive Environmental Stresses. Cells, 8(11), 1307. https://doi.org/10.3390/cells8111307