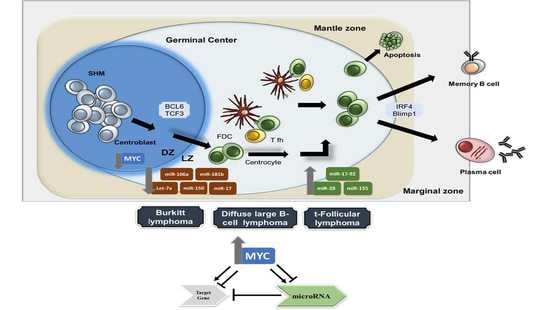
Germinal Centre B Cell Functions and Lymphomagenesis: Circuits Involving MYC and MicroRNAs
Abstract
:1. Introduction
2. Transcriptional Networks Involved in Physiological B-Cell Differentiation
3. MiRNAs in Germinal Centre B-Cell Functions and Lymphomagenesis
4. Germinal Centre-Derived B Cell Lymphomas with Abnormalities Involving MYC
4.1. Burkitt Lymphoma
4.2. Diffuse Large B-Cell Lymphoma
4.3. Follicular Lymphoma
5. MYC Feedback Loops Involving microRNAs and Their Roles in B-Cell Lymphomas
6. Predicted Models of FFLs among MYC, Its Targets, and miRNAs in Germinal Centre B-Cell Lymphomas
6.1. MYC-miR-17, miR-20-E2F1 (Type 1 FFL Circuit)
6.2. MYC-miR-29a/b/c-DNMT3B (Type 2 FFL Circuit)
6.3. MYC–miR-19a–PTEN (Type 1 or 2 FFL Circuit)
6.4. MYC–miR-150–FOXP1
7. Remarks and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dang, C.V.; O’Donnell, K.A.; Zeller, K.I.; Nguyen, T.; Osthus, R.C.; Li, F. The c-Myc target gene network. Semin. Cancer Biol. 2006, 16, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Menssen, A.; Hermeking, H. Characterization of the c-MYC-regulated transcriptome by SAGE: Identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. USA 2002, 99, 6274–6279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amati, B.; Brooks, M.W.; Levy, N.; Littlewood, T.D.; Evan, G.I.; Land, H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 1993, 72, 233–245. [Google Scholar] [CrossRef]
- Dalla-Favera, R.; Bregni, M.; Erikson, J.; Patterson, D.; Gallo, R.C.; Croce, C.M. Human c-Myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7824–7827. [Google Scholar] [CrossRef] [PubMed]
- Taub, R.; Kirsch, I.; Morton, C.; Lenoir, G.; Swan, D.; Tronick, S.; Aaronson, S.; Leder, P. Translocation of the c-Myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7837–7841. [Google Scholar] [CrossRef] [PubMed]
- Shou, Y.; Martelli, M.L.; Gabrea, A.; Qi, Y.; Brents, L.A.; Roschke, A.; Dewald, G.; Kirsch, I.R.; Bergsagel, P.L.; Kuehl, W.M. Diverse karyotypic abnormalities of the c-Myc locus associated with c-Myc dysregulation and tumor progression in multiple myeloma. Proc. Natl. Acad. Sci. USA 2000, 97, 228–233. [Google Scholar] [CrossRef]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef]
- Salghetti, S.E.; Kim, S.Y.; Tansey, W.P. Destruction of Myc by ubiquitin-mediated proteolysis: Cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999, 18, 717–726. [Google Scholar] [CrossRef]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a target of the APC pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef]
- Gregory, M.A.; Hann, S.R. c-Myc proteolysis by the ubiquitin-proteasome pathway: Stabilization of c-Myc in Burkitt’s lymphoma cells. Mol. Cell Biol 2000, 20, 2423–2435. [Google Scholar] [CrossRef]
- King, B.; Trimarchi, T.; Reavie, L.; Xu, L.; Mullenders, J.; Ntziachristos, P.; Aranda-Orgilles, B.; Perez-Garcia, A.; Shi, J.; Vakoc, C.; et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 2013, 153, 1552–1566. [Google Scholar] [CrossRef] [PubMed]
- Jackstadt, R.; Menssen, A.; Hermeking, H. Genome-wide analysis of c-MYC-regulated mRNAs and miRNAs, and c-MYC DNA binding by next-generation sequencing. Methods Mol. Biol 2013, 1012, 145–185. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Moutinho, C.; Esteller, M. MicroRNAs and Epigenetics. Adv. Cancer Res. 2017, 135, 189–220. [Google Scholar] [CrossRef]
- Psathas, J.N.; Thomas-Tikhonenko, A. MYC and the art of microRNA maintenance. Cold Spring Harb. Perspect. Med. 2014, 4. [Google Scholar] [CrossRef]
- Xiong, L.; Jiang, W.; Zhou, R.; Mao, C.; Guo, Z. Identification and analysis of the regulatory network of Myc and microRNAs from high-throughput experimental data. Comput. Biol. Med. 2013, 43, 1252–1260. [Google Scholar] [CrossRef]
- Milo, R.; Shen-Orr, S.; Itzkovitz, S.; Kashtan, N.; Chklovskii, D.; Alon, U. Network motifs: Simple building blocks of complex networks. Science 2002, 298, 824–827. [Google Scholar] [CrossRef]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839. [Google Scholar] [CrossRef] [PubMed]
- Herold, S.; Wanzel, M.; Beuger, V.; Frohme, C.; Beul, D.; Hillukkala, T.; Syvaoja, J.; Saluz, H.P.; Haenel, F.; Eilers, M. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell 2002, 10, 509–521. [Google Scholar] [CrossRef]
- Kleine-Kohlbrecher, D.; Adhikary, S.; Eilers, M. Mechanisms of transcriptional repression by Myc. Curr. Top. Microbiol. Immunol. 2006, 302, 51–62. [Google Scholar] [PubMed]
- Schmitz, U.; Wolkenhauer, O.; Vera, J. MicroRNA Cancer Regulation: Advanced Concepts, Bioinformatics and Systems Biology Tools; Springer Science & Business Media: Berlin, Germany, 2013; Volume 774. [Google Scholar]
- Bueno, M.J.; Gomez de Cedron, M.; Gomez-Lopez, G.; Perez de Castro, I.; Di Lisio, L.; Montes-Moreno, S.; Martinez, N.; Guerrero, M.; Sanchez-Martinez, R.; Santos, J.; et al. Combinatorial effects of microRNAs to suppress the Myc oncogenic pathway. Blood 2011, 117, 6255–6266. [Google Scholar] [CrossRef]
- Sotillo, E.; Laver, T.; Mellert, H.; Schelter, J.M.; Cleary, M.A.; McMahon, S.; Thomas-Tikhonenko, A. Myc overexpression brings out unexpected antiapoptotic effects of miR-34a. Oncogene 2011, 30, 2587–2594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannell, I.; Bushell, M. Regulation of Myc by miR-34c: A mechanism to prevent genomic instability? Cell Cycle 2014, 9, 2798–2802. [Google Scholar] [CrossRef]
- Sampson, V.B.; Rong, N.H.; Han, J.; Yang, Q.; Aris, V.; Soteropoulos, P.; Petrelli, N.J.; Dunn, S.P.; Krueger, L.J. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007, 67, 9762–9770. [Google Scholar] [CrossRef]
- Jackstadt, R.; Hermeking, H. MicroRNAs as regulators and mediators of c-MYC function. Biochim. Biophys. Acta 2015, 1849, 544–553. [Google Scholar] [CrossRef]
- Zhao, X.; Lwin, T.; Zhang, X.; Huang, A.; Wang, J.; Marquez, V.E.; Chen-Kiang, S.; Dalton, W.S.; Sotomayor, E.; Tao, J. Disruption of the MYC-miRNA-EZH2 loop to suppress aggressive B-cell lymphoma survival and clonogenicity. Leukemia 2013, 27, 2341–2350. [Google Scholar] [CrossRef] [Green Version]
- Bui, T.V.; Mendell, J.T. Myc: Maestro of MicroRNAs. Genes Cancer 2010, 1, 568–575. [Google Scholar] [CrossRef]
- Rajewsky, K. Clonal selection and learning in the antibody system. Nature 1996, 381, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Medina, K.L.; Singh, H. Genetic networks that regulate B lymphopoiesis. Curr. Opin. Hematol. 2005, 12, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Nutt, S.L.; Kee, B.L. The transcriptional regulation of B cell lineage commitment. Immunity 2007, 26, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Herzog, S.; Reth, M.; Jumaa, H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat. Rev. Immunol. 2009, 9, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Mandel, E.M.; Grosschedl, R. Transcription control of early B cell differentiation. Curr. Opin. Immunol. 2010, 22, 161–167. [Google Scholar] [CrossRef]
- De Silva, N.S.; Klein, U. Dynamics of B cells in germinal centres. Nat. Rev. Immunol. 2015, 15, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Levy, N.S.; Malipiero, U.V.; Lebecque, S.G.; Gearhart, P.J. Early onset of somatic mutation in immunoglobulin VH genes during the primary immune response. J. Exp. Med. 1989, 169, 2007–2019. [Google Scholar] [CrossRef]
- Honjo, T.; Kinoshita, K.; Muramatsu, M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu. Rev. Immunol 2002, 20, 165–196. [Google Scholar] [CrossRef]
- Berek, C.; Berger, A.; Apel, M. Maturation of the immune response in germinal centers. Cell 1991, 67, 1121–1129. [Google Scholar] [CrossRef]
- Jacob, J.; Kelsoe, G.; Rajewsky, K.; Weiss, U. Intraclonal generation of antibody mutants in germinal centres. Nature 1991, 354, 389–392. [Google Scholar] [CrossRef]
- Victora, G.D.; Dominguez-Sola, D.; Holmes, A.B.; Deroubaix, S.; Dalla-Favera, R.; Nussenzweig, M.C. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood 2012, 120, 2240–2248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basso, K.; Dalla-Favera, R. Germinal centres and B cell lymphomagenesis. Nat. Rev. Immunol. 2015, 15, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Basso, K.; Schneider, C.; Shen, Q.; Holmes, A.B.; Setty, M.; Leslie, C.; Dalla-Favera, R. BCL6 positively regulates AID and germinal center gene expression via repression of miR-155. J. Exp. Med. 2012, 209, 2455–2465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, U.; Tu, Y.; Stolovitzky, G.A.; Keller, J.L.; Haddad, J., Jr.; Miljkovic, V.; Cattoretti, G.; Califano, A.; Dalla-Favera, R. Transcriptional analysis of the B cell germinal center reaction. Proc. Natl. Acad. Sci. USA 2003, 100, 2639–2644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ci, W.; Polo, J.M.; Cerchietti, L.; Shaknovich, R.; Wang, L.; Yang, S.N.; Ye, K.; Farinha, P.; Horsman, D.E.; Gascoyne, R.D.; et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood 2009, 113, 5536–5548. [Google Scholar] [CrossRef] [Green Version]
- Basso, K.; Saito, M.; Sumazin, P.; Margolin, A.A.; Wang, K.; Lim, W.K.; Kitagawa, Y.; Schneider, C.; Alvarez, M.J.; Califano, A.; et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood 2010, 115, 975–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez-Sola, D.; Victora, G.D.; Ying, C.Y.; Phan, R.T.; Saito, M.; Nussenzweig, M.C.; Dalla-Favera, R. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat. Immunol. 2012, 13, 1083. [Google Scholar] [CrossRef]
- Calado, D.P.; Sasaki, Y.; Godinho, S.A.; Pellerin, A.; Köchert, K.; Sleckman, B.P.; de Alborán, I.M.; Janz, M.; Rodig, S.; Rajewsky, K. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat. Immun. 2012, 13, 1092. [Google Scholar] [CrossRef]
- Klein, U.; Dalla-Favera, R. Germinal centres: Role in B-cell physiology and malignancy. Nat. Rev. Immunol. 2008, 8, 22–33. [Google Scholar] [CrossRef]
- Danger, R.; Braza, F.; Giral, M.; Soulillou, J.P.; Brouard, S. MicroRNAs, Major Players in B Cells Homeostasis and Function. Front. Immunol. 2014, 5, 98. [Google Scholar] [CrossRef]
- Coffre, M.; Koralov, S.B. miRNAs in B Cell Development and Lymphomagenesis. Trends Mol. Med. 2017, 23, 721–736. [Google Scholar] [CrossRef] [PubMed]
- Koralov, S.B.; Muljo, S.A.; Galler, G.R.; Krek, A.; Chakraborty, T.; Kanellopoulou, C.; Jensen, K.; Cobb, B.S.; Merkenschlager, M.; Rajewsky, N.; et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 2008, 132, 860–874. [Google Scholar] [CrossRef] [PubMed]
- Coffre, M.; Benhamou, D.; Riess, D.; Blumenberg, L.; Snetkova, V.; Hines, M.J.; Chakraborty, T.; Bajwa, S.; Jensen, K.; Chong, M.M.W.; et al. miRNAs Are Essential for the Regulation of the PI3K/AKT/FOXO Pathway and Receptor Editing during B Cell Maturation. Cell Rep. 2016, 17, 2271–2285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.P.; Wang, M.; Robertus, J.L.; Schakel, R.N.; Gibcus, J.H.; Diepstra, A.; Harms, G.; Peh, S.C.; Reijmers, R.M.; Pals, S.T.; et al. miRNA profiling of B-cell subsets: Specific miRNA profile for germinal center B cells with variation between centroblasts and centrocytes. Lab. Investig. 2009, 89, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Basso, K.; Sumazin, P.; Morozov, P.; Schneider, C.; Maute, R.L.; Kitagawa, Y.; Mandelbaum, J.; Haddad, J., Jr.; Chen, C.Z.; Califano, A.; et al. Identification of the human mature B cell miRNome. Immunity 2009, 30, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Ridzon, D.; Wong, L.; Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genom. 2007, 8, 166. [Google Scholar] [CrossRef]
- Monticelli, S.; Ansel, K.M.; Xiao, C.; Socci, N.D.; Krichevsky, A.M.; Thai, T.H.; Rajewsky, N.; Marks, D.S.; Sander, C.; Rajewsky, K.; et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005, 6, R71. [Google Scholar] [CrossRef]
- Schneider, C.; Setty, M.; Holmes, A.B.; Maute, R.L.; Leslie, C.S.; Mussolin, L.; Rosolen, A.; Dalla-Favera, R.; Basso, K. MicroRNA 28 controls cell proliferation and is down-regulated in B-cell lymphomas. Proc. Natl. Acad. Sci. USA 2014, 111, 8185–8190. [Google Scholar] [CrossRef]
- Olive, V.; Li, Q.; He, L. mir-17-92: A polycistronic oncomir with pleiotropic functions. Immunol. Rev. 2013, 253, 158–166. [Google Scholar] [CrossRef]
- Xu, S.; Guo, K.; Zeng, Q.; Huo, J.; Lam, K.P. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood 2012, 119, 767–776. [Google Scholar] [CrossRef]
- Thai, T.H.; Calado, D.P.; Casola, S.; Ansel, K.M.; Xiao, C.; Xue, Y.; Murphy, A.; Frendewey, D.; Valenzuela, D.; Kutok, J.L.; et al. Regulation of the germinal center response by microRNA-155. Science 2007, 316, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Vigorito, E.; Perks, K.L.; Abreu-Goodger, C.; Bunting, S.; Xiang, Z.; Kohlhaas, S.; Das, P.P.; Miska, E.A.; Rodriguez, A.; Bradley, A.; et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007, 27, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Kluiver, J.; Haralambieva, E.; de Jong, D.; Blokzijl, T.; Jacobs, S.; Kroesen, B.J.; Poppema, S.; van den Berg, A. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosom. Cancer 2006, 45, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Robertus, J.L.; Kluiver, J.; Weggemans, C.; Harms, G.; Reijmers, R.M.; Swart, Y.; Kok, K.; Rosati, S.; Schuuring, E.; van Imhoff, G.; et al. MiRNA profiling in B non-Hodgkin lymphoma: A MYC-related miRNA profile characterizes Burkitt lymphoma. Br. J. Haematol. 2010, 149, 896–899. [Google Scholar] [CrossRef] [PubMed]
- Kluiver, J.; van den Berg, A.; de Jong, D.; Blokzijl, T.; Harms, G.; Bouwman, E.; Jacobs, S.; Poppema, S.; Kroesen, B.J. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene 2007, 26, 3769–3776. [Google Scholar] [CrossRef] [PubMed]
- De Yebenes, V.G.; Bartolome-Izquierdo, N.; Ramiro, A.R. Regulation of B-cell development and function by microRNAs. Immunol. Rev. 2013, 253, 25–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramiro, A.R.; Jankovic, M.; Eisenreich, T.; Difilippantonio, S.; Chen-Kiang, S.; Muramatsu, M.; Honjo, T.; Nussenzweig, A.; Nussenzweig, M.C. AID is required for c-Myc/IgH chromosome translocations in vivo. Cell 2004, 118, 431–438. [Google Scholar] [CrossRef]
- Ramiro, A.R.; Jankovic, M.; Callen, E.; Difilippantonio, S.; Chen, H.T.; McBride, K.M.; Eisenreich, T.R.; Chen, J.; Dickins, R.A.; Lowe, S.W.; et al. Role of genomic instability and p53 in AID-induced c-Myc-Igh translocations. Nature 2006, 440, 105–109. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Bothmer, A.; Callen, E.; Reina-San-Martin, B.; Dorsett, Y.; Difilippantonio, S.; Bolland, D.J.; Chen, H.T.; Corcoran, A.E.; Nussenzweig, A.; et al. AID is required for the chromosomal breaks in c-Myc that lead to c-Myc/IgH translocations. Cell 2008, 135, 1028–1038. [Google Scholar] [CrossRef]
- Teng, G.; Hakimpour, P.; Landgraf, P.; Rice, A.; Tuschl, T.; Casellas, R.; Papavasiliou, F.N. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity 2008, 28, 621–629. [Google Scholar] [CrossRef]
- Dorsett, Y.; McBride, K.M.; Jankovic, M.; Gazumyan, A.; Thai, T.H.; Robbiani, D.F.; Di Virgilio, M.; Reina San-Martin, B.; Heidkamp, G.; Schwickert, T.A.; et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity 2008, 28, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Eis, P.S.; Tam, W.; Sun, L.; Chadburn, A.; Li, Z.; Gomez, M.F.; Lund, E.; Dahlberg, J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 2005, 102, 3627–3632. [Google Scholar] [CrossRef] [PubMed]
- Pasqualucci, L.; Bhagat, G.; Jankovic, M.; Compagno, M.; Smith, P.; Muramatsu, M.; Honjo, T.; Morse, H.C., III; Nussenzweig, M.C.; Dalla-Favera, R. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 2008, 40, 108–112. [Google Scholar] [CrossRef] [PubMed]
- De Yebenes, V.G.; Belver, L.; Pisano, D.G.; Gonzalez, S.; Villasante, A.; Croce, C.; He, L.; Ramiro, A.R. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med. 2008, 205, 2199–2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Wan, Y.; Ji, Q.; Fang, Y.; Wu, Y. The role of microRNAs in B-cell development and function. Cell Mol. Immunol. 2013, 10, 107–112. [Google Scholar] [CrossRef] [Green Version]
- Kuchen, S.; Resch, W.; Yamane, A.; Kuo, N.; Li, Z.; Chakraborty, T.; Wei, L.; Laurence, A.; Yasuda, T.; Peng, S.; et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 2010, 32, 828–839. [Google Scholar] [CrossRef]
- Zheng, B.; Xi, Z.; Liu, R.; Yin, W.; Sui, Z.; Ren, B.; Miller, H.; Gong, Q.; Liu, C. The Function of MicroRNAs in B-Cell Development, Lymphoma, and Their Potential in Clinical Practice. Front. Immunol. 2018, 9, 936. [Google Scholar] [CrossRef]
- Robaina, M.C.; Faccion, R.S.; Mazzoccoli, L.; Rezende, L.M.M.; Queiroga, E.; Bacchi, C.E.; Thomas-Tikhonenko, A.; Klumb, C.E. miR-17-92 cluster components analysis in Burkitt lymphoma: Overexpression of miR-17 is associated with poor prognosis. Ann. Hematol. 2016, 95, 881–891. [Google Scholar] [CrossRef]
- Robaina, M.C.; Mazzoccoli, L.; Arruda, V.O.; Reis, F.R.; Apa, A.G.; de Rezende, L.M.; Klumb, C.E. Deregulation of DNMT1, DNMT3B and miR-29s in Burkitt lymphoma suggests novel contribution for disease pathogenesis. Exp. Mol. Pathol 2015, 98, 200–207. [Google Scholar] [CrossRef]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704. [Google Scholar] [CrossRef]
- Nguyen, L.; Papenhausen, P.; Shao, H. The role of c-MYC in B-cell lymphomas: Diagnostic and molecular aspects. Genes 2017, 8, 116. [Google Scholar] [CrossRef] [PubMed]
- Korac, P.; Dotlic, S.; Matulic, M.; Zajc Petranovic, M.; Dominis, M. Role of MYC in B Cell Lymphomagenesis. Genes 2017, 8, 115. [Google Scholar] [CrossRef] [PubMed]
- Wagener, R.; López, C.; Siebert, R. Pathogenesis of B-Cell Lymphoma. In Non-Hodgkin’s Lymphoma in Childhood and Adolescence; Springer: Berlin, Germany, 2019; pp. 33–50. [Google Scholar]
- Grimm, K.E.; O’Malley, D.P. Aggressive B cell lymphomas in the 2017 revised WHO classification of tumors of hematopoietic and lymphoid tissues. Ann. Diagn. Pathol. 2019, 38, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Cory, S. Activation of cellular oncogenes in hemopoietic cells by chromosome translocation. Adv. Cancer Res. 1986, 47, 189–234. [Google Scholar]
- Slack, G.W.; Gascoyne, R.D. MYC and aggressive B-cell lymphomas. Adv. Anat. Pathol. 2011, 18, 219–228. [Google Scholar] [CrossRef]
- Bemark, M.; Neuberger, M.S. The c-MYC allele that is translocated into the IgH locus undergoes constitutive hypermutation in a Burkitt’s lymphoma line. Oncogene 2000, 19, 3404–3410. [Google Scholar] [CrossRef]
- Gregory, M.A.; Xiao, Q.; Cornwall, G.A.; Lutterbach, B.; Hann, S.R. B-Myc is preferentially expressed in hormonally-controlled tissues and inhibits cellular proliferation. Oncogene 2000, 19, 4886–4895. [Google Scholar] [CrossRef] [Green Version]
- Sander, S.; Calado, D.P.; Srinivasan, L.; Kochert, K.; Zhang, B.; Rosolowski, M.; Rodig, S.J.; Holzmann, K.; Stilgenbauer, S.; Siebert, R.; et al. Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell 2012, 22, 167–179. [Google Scholar] [CrossRef]
- Love, C.; Sun, Z.; Jima, D.; Li, G.; Zhang, J.; Miles, R.; Richards, K.L.; Dunphy, C.H.; Choi, W.W.; Srivastava, G.; et al. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 2012, 44, 1321–1325. [Google Scholar] [CrossRef] [Green Version]
- Richter, J.; Schlesner, M.; Hoffmann, S.; Kreuz, M.; Leich, E.; Burkhardt, B.; Rosolowski, M.; Ammerpohl, O.; Wagener, R.; Bernhart, S.H.; et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat. Genet. 2012, 44, 1316–1320. [Google Scholar] [CrossRef]
- Tomita, N. BCL2 and MYC dual-hit lymphoma/leukemia. J. Clin. Exp. Hematopathol. 2011, 51, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Savage, K.J.; Johnson, N.A.; Ben-Neriah, S.; Connors, J.M.; Sehn, L.H.; Farinha, P.; Horsman, D.E.; Gascoyne, R.D. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood 2009, 114, 3533–3537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrans, S.; Crouch, S.; Smith, A.; Turner, K.; Owen, R.; Patmore, R.; Roman, E.; Jack, A. Rearrangement of MYC is associated with poor prognosis in patients with diffuse large B-cell lymphoma treated in the era of rituximab. J. Clin. Oncol. 2010, 28, 3360–3365. [Google Scholar] [CrossRef] [PubMed]
- Karube, K.; Campo, E. MYC alterations in diffuse large B-cell lymphomas. Semin. Hematol. 2015, 52, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Hummel, M.; Bentink, S.; Berger, H.; Klapper, W.; Wessendorf, S.; Barth, T.F.; Bernd, H.W.; Cogliatti, S.B.; Dierlamm, J.; Feller, A.C.; et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N. Engl. J. Med. 2006, 354, 2419–2430. [Google Scholar] [CrossRef]
- Pasqualucci, L. Molecular pathogenesis of germinal center-derived B cell lymphomas. Immunol. Rev. 2019, 288, 240–261. [Google Scholar] [CrossRef]
- Valera, A.; Lopez-Guillermo, A.; Cardesa-Salzmann, T.; Climent, F.; Gonzalez-Barca, E.; Mercadal, S.; Espinosa, I.; Novelli, S.; Briones, J.; Mate, J.L.; et al. MYC protein expression and genetic alterations have prognostic impact in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Haematologica 2013, 98, 1554–1562. [Google Scholar] [CrossRef]
- Cai, Q.; Medeiros, L.J.; Xu, X.; Young, K.H. MYC-driven aggressive B-cell lymphomas: Biology, entity, differential diagnosis and clinical management. Oncotarget 2015, 6, 38591–38616. [Google Scholar] [CrossRef]
- Schraders, M.; de Jong, D.; Kluin, P.; Groenen, P.; van Krieken, H. Lack of Bcl-2 expression in follicular lymphoma may be caused by mutations in the BCL2 gene or by absence of the t (14;18) translocation. J. Pathol. 2005, 205, 329–335. [Google Scholar] [CrossRef]
- Yano, T.; Jaffe, E.S.; Longo, D.L.; Raffeld, M. MYC rearrangements in histologically progressed follicular lymphomas. Blood 1992, 80, 758–767. [Google Scholar] [CrossRef] [Green Version]
- Montoto, S.; Davies, A.J.; Matthews, J.; Calaminici, M.; Norton, A.J.; Amess, J.; Vinnicombe, S.; Waters, R.; Rohatiner, A.Z.; Lister, T.A. Risk and clinical implications of transformation of follicular lymphoma to diffuse large B-cell lymphoma. J. Clin. Oncol. 2007, 25, 2426–2433. [Google Scholar] [CrossRef] [PubMed]
- Okosun, J.; Bodor, C.; Wang, J.; Araf, S.; Yang, C.Y.; Pan, C.; Boller, S.; Cittaro, D.; Bozek, M.; Iqbal, S.; et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014, 46, 176–181. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Khiabanian, H.; Fangazio, M.; Vasishtha, M.; Messina, M.; Holmes, A.B.; Ouillette, P.; Trifonov, V.; Rossi, D.; Tabbo, F.; et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014, 6, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Kridel, R.; Mottok, A.; Farinha, P.; Ben-Neriah, S.; Ennishi, D.; Zheng, Y.; Chavez, E.A.; Shulha, H.P.; Tan, K.; Chan, F.C.; et al. Cell of origin of transformed follicular lymphoma. Blood 2015, 126, 2118–2127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, A.J.; Rosenwald, A.; Wright, G.; Lee, A.; Last, K.W.; Weisenburger, D.D.; Chan, W.C.; Delabie, J.; Braziel, R.M.; Campo, E.; et al. Transformation of follicular lymphoma to diffuse large B-cell lymphoma proceeds by distinct oncogenic mechanisms. Br. J. Haematol. 2007, 136, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Fischer, T.; Zing, N.P.C.; Chiattone, C.S.; Federico, M.; Luminari, S. Transformed follicular lymphoma. Ann. Hematol. 2018, 97, 17–29. [Google Scholar] [CrossRef]
- Mangan, S.; Alon, U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 2003, 100, 11980–11985. [Google Scholar] [CrossRef] [Green Version]
- Kress, T.R.; Sabo, A.; Amati, B. MYC: Connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer 2015, 15, 593–607. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA: RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Tsang, J.; Zhu, J.; van Oudenaarden, A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 2007, 26, 753–767. [Google Scholar] [CrossRef]
- Zhang, H.M.; Kuang, S.; Xiong, X.; Gao, T.; Liu, C.; Guo, A.Y. Transcription factor and microRNA co-regulatory loops: Important regulatory motifs in biological processes and diseases. Brief. Bioinf. 2015, 16, 45–58. [Google Scholar] [CrossRef]
- El Baroudi, M.; Cora, D.; Bosia, C.; Osella, M.; Caselle, M. A curated database of miRNA mediated feed-forward loops involving MYC as master regulator. PLoS ONE 2011, 6, e14742. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ma, C.; Xing, C.; Chen, C.L.; Chen, Z.; Yao, Y.; Wang, J.; Tao, C. Burkitt lymphoma-associated network construction and important network motif analysis. Oncol. Lett. 2018, 16, 3054–3062. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Alidadiani, N.; Ghaderi, S.; Dilaver, N.; Bakhshamin, S.; Bayat, M. Epithelial mesenchymal transition Transcription Factor (TF): The structure, function and microRNA feedback loop. Gene 2018, 674, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Zeller, K.I.; Jegga, A.G.; Aronow, B.J.; O’Donnell, K.A.; Dang, C.V. An integrated database of genes responsive to the Myc oncogenic transcription factor: Identification of direct genomic targets. Genome Biol. 2003, 4, R69. [Google Scholar] [CrossRef]
- Wang, J.; Lu, M.; Qiu, C.; Cui, Q. TransmiR: A transcription factor-microRNA regulation database. Nucleic Acids Res. 2010, 38, D119–D122. [Google Scholar] [CrossRef]
- Papadopoulos, G.L.; Reczko, M.; Simossis, V.A.; Sethupathy, P.; Hatzigeorgiou, A.G. The database of experimentally supported targets: A functional update of TarBase. Nucleic Acids Res. 2009, 37, D155–D158. [Google Scholar] [CrossRef]
- Iqbal, J.; Shen, Y.; Huang, X.; Liu, Y.; Wake, L.; Liu, C.; Deffenbacher, K.; Lachel, C.M.; Wang, C.; Rohr, J.; et al. Global microRNA expression profiling uncovers molecular markers for classification and prognosis in aggressive B-cell lymphoma. Blood 2015, 125, 1137–1145. [Google Scholar] [CrossRef]
- Dave, S.S.; Fu, K.; Wright, G.W.; Lam, L.T.; Kluin, P.; Boerma, E.J.; Greiner, T.C.; Weisenburger, D.D.; Rosenwald, A.; Ott, G.; et al. Molecular diagnosis of Burkitt’s lymphoma. N. Engl. J. Med. 2006, 354, 2431–2442. [Google Scholar] [CrossRef]
- Chang, T.C.; Yu, D.; Lee, Y.S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008, 40, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Malpeli, G.; Barbi, S.; Tosadori, G.; Greco, C.; Zupo, S.; Pedron, S.; Brunelli, M.; Bertolaso, A.; Scupoli, M.T.; Krampera, M.; et al. MYC-related microRNAs signatures in non-Hodgkin B-cell lymphomas and their relationships with core cellular pathways. Oncotarget 2018, 9, 29753–29771. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.P.; Ciro, M.; Cocito, A.; Helin, K. E2F target genes: Unraveling the biology. Trends Biochem. Sci. 2004, 29, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Hallstrom, T.C.; Mori, S.; Nevins, J.R. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell 2008, 13, 11–22. [Google Scholar] [CrossRef]
- Emmrich, S.; Putzer, B.M. Checks and balances: E2F-microRNA crosstalk in cancer control. Cell Cycle 2010, 9, 2555–2567. [Google Scholar] [CrossRef]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef]
- McMahon, S.B.; Van Buskirk, H.A.; Dugan, K.A.; Copeland, T.D.; Cole, M.D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 1998, 94, 363–374. [Google Scholar] [CrossRef]
- Wang, H.; Larris, B.; Peiris, T.H.; Zhang, L.; Le Lay, J.; Gao, Y.; Greenbaum, L.E. C/EBPbeta activates E2F-regulated genes in vivo via recruitment of the coactivator CREB-binding protein/P300. J. Biol. Chem. 2007, 282, 24679–24688. [Google Scholar] [CrossRef]
- Zeller, K.I.; Zhao, X.; Lee, C.W.; Chiu, K.P.; Yao, F.; Yustein, J.T.; Ooi, H.S.; Orlov, Y.L.; Shahab, A.; Yong, H.C.; et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl. Acad. Sci. USA 2006, 103, 17834–17839. [Google Scholar] [CrossRef] [Green Version]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Ota, A.; Tagawa, H.; Karnan, S.; Tsuzuki, S.; Karpas, A.; Kira, S.; Yoshida, Y.; Seto, M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004, 64, 3087–3095. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, P.C.; Frank, S.R.; Wang, L.; Schroeder, M.; Liu, S.; Greene, J.; Cocito, A.; Amati, B. Genomic targets of the human c-Myc protein. Genes Dev. 2003, 17, 1115–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, J.Y.; Ehmann, G.L.; Giangrande, P.H.; Nevins, J.R. A role for Myc in facilitating transcription activation by E2F1. Oncogene 2008, 27, 4172–4179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumura, I.; Tanaka, H.; Kanakura, Y. E2F1 and c-Myc in cell growth and death. Cell Cycle 2003, 2, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Coller, H.A.; Forman, J.J.; Legesse-Miller, A. “Myc’ed message”: Myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet. 2007, 3, e146. [Google Scholar] [CrossRef]
- Molina-Privado, I.; Rodriguez-Martinez, M.; Rebollo, P.; Martin-Perez, D.; Artiga, M.J.; Menarguez, J.; Flemington, E.K.; Piris, M.A.; Campanero, M.R. E2F1 expression is deregulated and plays an oncogenic role in sporadic Burkitt’s lymphoma. Cancer Res. 2009, 69, 4052–4058. [Google Scholar] [CrossRef]
- Fabbri, M.; Ivan, M.; Cimmino, A.; Negrini, M.; Calin, G.A. Regulatory mechanisms of microRNAs involvement in cancer. Expert Opin. Biol. Ther. 2007, 7, 1009–1019. [Google Scholar] [CrossRef]
- Zhao, J.J.; Lin, J.; Lwin, T.; Yang, H.; Guo, J.; Kong, W.; Dessureault, S.; Moscinski, L.C.; Rezania, D.; Dalton, W.S.; et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood 2010, 115, 2630–2639. [Google Scholar] [CrossRef]
- Kinoshita, T.; Nohata, N.; Hanazawa, T.; Kikkawa, N.; Yamamoto, N.; Yoshino, H.; Itesako, T.; Enokida, H.; Nakagawa, M.; Okamoto, Y. Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin–integrin signalling in head and neck squamous cell carcinoma. Br. J. Cancer 2013, 109, 2636–2645. [Google Scholar] [CrossRef]
- Amodio, N.; Rossi, M.; Raimondi, L.; Pitari, M.R.; Botta, C.; Tagliaferri, P.; Tassone, P. miR-29s: A family of epi-miRNAs with therapeutic implications in hematologic malignancies. Oncotarget 2015, 6, 12837–12861. [Google Scholar] [CrossRef]
- Mott, J.L.; Kurita, S.; Cazanave, S.C.; Bronk, S.F.; Werneburg, N.W.; Fernandez-Zapico, M.E. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-κB. J. Cell Biochem. 2010, 110, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Garzon, R.; Sun, H.; Ladner, K.J.; Singh, R.; Dahlman, J.; Cheng, A.; Hall, B.M.; Qualman, S.J.; Chandler, D.S.; et al. NF-κB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 2008, 14, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, X.; Fiskus, W.; Lin, J.; Lwin, T.; Rao, R.; Zhang, Y.; Chan, J.C.; Fu, K.; Marquez, V.E.; et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-cell lymphomas. Cancer Cell 2012, 22, 506–523. [Google Scholar] [CrossRef] [PubMed]
- Mazzoccoli, L.; Robaina, M.C.; Apa, A.G.; Bonamino, M.; Pinto, L.W.; Queiroga, E.; Bacchi, C.E.; Klumb, C.E. MiR-29 silencing modulates the expression of target genes related to proliferation, apoptosis and methylation in Burkitt lymphoma cells. J. Cancer Res. Clin. Oncol. 2018, 144, 483–497. [Google Scholar] [CrossRef]
- Mazzoccoli, L.; Robaina, M.C.; Bacchi, C.E.; Soares Lima, S.C.; Klumb, C.E. miR-29 promoter and enhancer methylation identified by pyrosequencing in Burkitt lymhoma cells: Interplay between MYC and miR-29 regulation. Oncol. Rep. 2019, 42, 775–784. [Google Scholar] [CrossRef]
- Di Lisio, L.; Sanchez-Beato, M.; Gomez-Lopez, G.; Rodriguez, M.E.; Montes-Moreno, S.; Mollejo, M.; Menarguez, J.; Martinez, M.A.; Alves, F.J.; Pisano, D.G.; et al. MicroRNA signatures in B-cell lymphomas. Blood Cancer J. 2012, 2, e57. [Google Scholar] [CrossRef]
- Kwon, J.J.; Factora, T.D.; Dey, S.; Kota, J. A Systematic Review of miR-29 in Cancer. Mol. Ther. Oncolytics 2019, 12, 173–194. [Google Scholar] [CrossRef]
- Lenz, G.; Wright, G.W.; Emre, N.C.; Kohlhammer, H.; Dave, S.S.; Davis, R.E.; Carty, S.; Lam, L.T.; Shaffer, A.L.; Xiao, W.; et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl. Acad. Sci. USA 2008, 105, 13520–13525. [Google Scholar] [CrossRef] [Green Version]
- Sandhu, S.K.; Fassan, M.; Volinia, S.; Lovat, F.; Balatti, V.; Pekarsky, Y.; Croce, C.M. B-cell malignancies in microRNA Emu-miR-17~92 transgenic mice. Proc. Natl. Acad. Sci. USA 2013, 110, 18208–18213. [Google Scholar] [CrossRef]
- Tagawa, H.; Ikeda, S.; Sawada, K. Role of microRNA in the pathogenesis of malignant lymphoma. Cancer Sci. 2013, 104, 801–809. [Google Scholar] [CrossRef]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Olive, V.; Bennett, M.J.; Walker, J.C.; Ma, C.; Jiang, I.; Cordon-Cardo, C.; Li, Q.J.; Lowe, S.W.; Hannon, G.J.; He, L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009, 23, 2839–2849. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, M.; Loosveld, M.; Montpellier, B.; Navarro, J.M.; Quilichini, B.; Picard, C.; Di Cristofaro, J.; Bagnis, C.; Fossat, C.; Hernandez, L.; et al. Posttranscriptional deregulation of MYC via PTEN constitutes a major alternative pathway of MYC activation in T-cell acute lymphoblastic leukemia. Blood 2011, 117, 6650–6659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musilova, K.; Devan, J.; Cerna, K.; Seda, V.; Pavlasova, G.; Sharma, S.; Oppelt, J.; Pytlik, R.; Prochazka, V.; Prouzova, Z.; et al. miR-150 downregulation contributes to the high-grade transformation of follicular lymphoma by upregulating FOXP1 levels. Blood 2018, 132, 2389–2400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagardoy, A.; Martinez-Ferrandis, J.I.; Roa, S.; Bunting, K.L.; Aznar, M.A.; Elemento, O.; Shaknovich, R.; Fontan, L.; Fresquet, V.; Perez-Roger, I.; et al. Downregulation of FOXP1 is required during germinal center B-cell function. Blood 2013, 121, 4311–4320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerna, K.; Mraz, M. p53 limits B cell receptor (BCR) signalling: A new role for miR-34a and FOXP1. Oncotarget 2018, 9, 36409–36410. [Google Scholar] [CrossRef]




| miRNA | MYC-miRNA Effect | Target Gene | Cellular Pathways | MYC-Transcription Factor Effect | Type FFL* |
|---|---|---|---|---|---|
| hsa-miR-17 | Activation | E2F1 | regulation of apoptosis and cell cycle | Activation | T1a |
| Activation | VEGF | regulation of apoptosis and proliferation | Activation or Repression | T1a or T2a | |
| hsa-miR-19a | Activation | PTEN | regulation of apoptosis, cell cycle and proliferation | Activation or Repression | T1a or T2a |
| hsa-miR-20a | Activation | E2F1 | regulation of apoptosis and cell cycle | Activation | T1a |
| Activation | TGFBR2 | regulation of cell proliferation | Repression | T2a | |
| Activation | VEGF | regulation of apoptosis and proliferation | Activation or Repression | T1a or T2a | |
| hsa-miR-106a | Activation | RB1 | regulation of apoptosis, cell cycle and proliferation | Repression | T2a |
| Activation | VEGF | regulation of apoptosis and proliferation | Activation or Repression | T1a or T2a | |
| Activation | CDKN1A | regulation of apoptosis, cell cycle and proliferation | Repression | T2a | |
| hsa-miR-106b | Activation | CDKN1A | regulation of apoptosis, cell cycle and proliferation | Repression | T2a |
| Activation | E2F1 | regulation of apoptosis and cell cycle | Activation | T1a | |
| Activation | PTEN | regulation of apoptosis, cell cycle and proliferation | Activation or Repression | T1a or T2a | |
| Activation | VEGF | regulation of apoptosis and proliferation | Activation or Repression | T1a or T2a | |
| hsa-let-7a | Repression | NRAS | regulation of cell proliferation | Activation | T2b |
| Repression | CASP3 | induction of apoptosis | Repression | T1b | |
| hsa-miR-15a | Repression | VEGF | regulation of apoptosis and proliferation | Activation or Repression | T1b or T2b |
| hsa-miR-22 | Repression | PTEN | regulation of apoptosis, cell cycle and proliferation | Activation or Repression | T1b or T2b |
| hsa-miR-26a | Repression | PTEN | regulation of apoptosis, cell cycle and proliferation | Activation or Repression | T1b or T2b |
| hsa-miR-29a | Repression | DNMT3B | regulation of DNA methylation | Activation | T2b |
| hsa-miR-29b | Repression | DNMT3B | regulation of DNA methylation | Activation | T2b |
| hsa-miR-29c | Repression | DNMT3B | regulation of DNA methylation | Activation | T2b |
| hsa-miR-34a | Repression | CCND1 | regulation of cell proliferation and cell cycle | Repression | T1b |
| Repression | E2F3 | regulation of cell proliferation and cell cycle | Activation | T2b | |
| Repression | MYC | proliferation, cycle cell control, apoptosis, metabolism, and methylation | Repression | T1c |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robaina, M.C.; Mazzoccoli, L.; Esteves Klumb, C. Germinal Centre B Cell Functions and Lymphomagenesis: Circuits Involving MYC and MicroRNAs. Cells 2019, 8, 1365. https://doi.org/10.3390/cells8111365
Robaina MC, Mazzoccoli L, Esteves Klumb C. Germinal Centre B Cell Functions and Lymphomagenesis: Circuits Involving MYC and MicroRNAs. Cells. 2019; 8(11):1365. https://doi.org/10.3390/cells8111365
Chicago/Turabian StyleRobaina, Marcela Cristina, Luciano Mazzoccoli, and Claudete Esteves Klumb. 2019. "Germinal Centre B Cell Functions and Lymphomagenesis: Circuits Involving MYC and MicroRNAs" Cells 8, no. 11: 1365. https://doi.org/10.3390/cells8111365
APA StyleRobaina, M. C., Mazzoccoli, L., & Esteves Klumb, C. (2019). Germinal Centre B Cell Functions and Lymphomagenesis: Circuits Involving MYC and MicroRNAs. Cells, 8(11), 1365. https://doi.org/10.3390/cells8111365