Molecular and Functional Characterization of the Somatic PIWIL1/piRNA Pathway in Colorectal Cancer Cells
Abstract
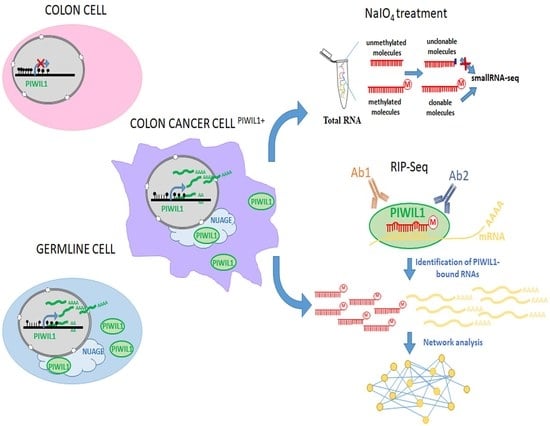
:1. Introduction
2. Materials and Methods
2.1. Gene Expression Databases and DNA Methylation Data Analysis
2.2. Cell Culture
2.3. Transient Transfections
2.4. Real-Time qRT-PCR
2.5. Total Protein Extraction
2.6. Cytosol/Nucleus Protein Fractionation
2.7. Cytosol/Perinucleus/Nucleus Protein Fractionation
2.8. SDS-Page and Western Blotting (WB)
2.9. Immunofluorescence (IF) Assay and Confocal Microscopy
2.10. Sodium-Periodate (NaIO4) Treatment/β-Elimination
2.11. RNA Extraction and Sequencing with Illumina Technologies
2.12. RNA Immunoprecipitation and Sequencing (RIP-Seq)
2.13. Bioinformatic Data Analysis
2.14. Availability of Supporting Data
3. Results
3.1. The Germline-Specific PIWIL1 Gene is Aberrantly Expressed in Colorectal Adenocarcinomas
3.2. PIWIL and piRNA Expression in CRC Cell Lines
3.3. Characterization of PIWI–piRNA Pathway in COLO 205
3.4. PIWIL1 Localizes in a Nuage-Like Structure of COLO 205 Cells
3.5. Identification of RNAs Associated with PIWIL1 in COLO 205 Cells
3.6. Identification of mRNAs Targeted by the PIWIL1–piRNA Complex
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Simpson, A.J.G.; Caballero, O.L.; Jungbluth, A.; Chen, Y.-T.; Old, L.J. Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer 2005, 5, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Gordeeva, O. Cancer-testis antigens: Unique cancer stem cell biomarkers and targets for cancer therapy. Semin. Cancer Boil. 2018, 53, 75–89. [Google Scholar] [CrossRef]
- Han, Y.-N.; Li, Y.; Xia, S.-Q.; Zhang, Y.-Y.; Zheng, J.-H.; Li, W. PIWI Proteins and PIWI-Interacting RNA: Emerging Roles in Cancer. Cell. Physiol. Biochem. 2017, 44, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojas-Ríos, P.; Simonelig, M. piRNAs and PIWI proteins: Regulators of gene expression in development and stem cells. Development 2018, 145, dev161786. [Google Scholar]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Song, L.; Liu, C.; Lv, X.; Li, X.; Jie, J.; Zhao, D.; Li, D. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumour Biol. 2016, 37, 2749–2756. [Google Scholar] [CrossRef] [PubMed]
- Rouget, C.; Papin, C.; Boureux, A.; Meunier, A.-C.; Franco, B.; Robine, N.; Lai, E.C.; Pélisson, A.; Simonelig, M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 2010, 467, 1128–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashim, A.; Rizzo, F.; Marchese, G.; Ravo, M.; Tarallo, R.; Nassa, G.; Giurato, G.; Santamaria, G.; Cordella, A.; Cantarella, C.; et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget 2014, 5, 9901–9910. [Google Scholar] [CrossRef]
- Martinez, V.D.; Vucic, E.A.; Thu, K.L.; Hubaux, R.; Enfield, K.S.; Pikor, L.A.; Becker-Santos, D.D.; Brown, C.J.; Lam, S.; Lam, W.L. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci. Rep. 2015, 5, 10423. [Google Scholar] [CrossRef]
- Ng, K.W.; Anderson, C.; Marshall, E.A.; Minatel, B.C.; Enfield, K.S.S.; Saprunoff, H.L.; Lam, W.L.; Martinez, V.D. Piwi-interacting RNAs in cancer: Emerging functions and clinical utility. Mol. Cancer 2016, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, F.; Rinaldi, A.; Marchese, G.; Coviellop, E.; Sellitto, A.; Cordella, A.; Giurato, G.; Nassa, G.; Ravo, M.; Tarallo, R.; et al. Specific patterns of PIWI-interacting small noncoding RNA expression in dysplastic liver nodules and hepatocellular carcinoma. Oncotarget 2016, 7, 54650–54661. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-J.; Kim, S.-M.; Kim, Y.-O.; Chang, H.-K. Clinicopathologic Implications of PIWIL2 Expression in Colorectal Cancer. Korean J. Pathol. 2012, 46, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Gao, Q.; Chen, K.; Xue, X.; Li, M.; Chen, Q.; Zhu, G.; Gao, Y. Hiwi facilitates chemoresistance as a cancer stem cell marker in cervical cancer. Oncol. Rep. 2014, 32, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Vychytilova-Faltejskova, P.; Stitkovcova, K.; Radova, L.; Sachlova, M.; Kosarova, Z.; Slaba, K.; Kala, Z.; Svoboda, M.; Kiss, I.; Vyzula, R.; et al. Circulating PIWI-Interacting RNAs piR-5937 and piR-28876 Are Promising Diagnostic Biomarkers of Colon Cancer. Cancer Epidemiol. Biomark. Prev. 2018, 27, 1019–1028. [Google Scholar] [CrossRef] [Green Version]
- Weng, M.; Wu, D.; Yang, C.; Peng, H.; Wang, G.; Wang, T.; Li, X. Noncoding RNAs in the development, diagnosis, and prognosis of colorectal cancer. Transl. Res. 2017, 181, 108–120. [Google Scholar] [CrossRef]
- Koduru, S.V.; Tiwari, A.K.; Hazard, S.W.; Mahajan, M.; Ravnic, D.J. Exploration of small RNA-seq data for small non-coding RNAs in Human Colorectal Cancer. Int. J. Genom. 2017, 5, 16–31. [Google Scholar] [CrossRef] [Green Version]
- Sun, R.; Gao, C.-L.; Li, D.-H.; Li, B.-J.; Ding, Y.-H. Expression Status of PIWIL1 as a Prognostic Marker of Colorectal Cancer. Dis. Markers 2017, 2017, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Raeisossadati, R.; Abbaszadegan, M.R.; Moghbeli, M.; Tavassoli, A.; Kihara, A.H.; Forghanifard, M.M. Aberrant expression of DPPA2 and HIWI genes in colorectal cancer and their impacts on poor prognosis. Tumour Biol. 2014, 35, 5299–5305. [Google Scholar] [CrossRef]
- Wang, H.L.; Chen, B.B.; Cao, X.G.; Wang, J.; Hu, X.F.; Mu, X.Q.; Chen, X.B. The clinical significances of the abnormal expressions of Piwil1 and Piwil2 in colonic adenoma and adenocarcinoma. Onco. Targets Ther. 2015, 8, 1259–1264. [Google Scholar] [CrossRef]
- Zeng, Y.; Qu, L.-K.; Meng, L.; Liu, C.-Y.; Dong, B.; Xing, X.-F.; Wu, J.; Shou, C.-C. HIWI expression profile in cancer cells and its prognostic value for patients with colorectal cancer. Chin. Med. J. 2011, 124, 2144–2149. [Google Scholar]
- Litwin, M.; Dubis, J.; Arczynska, K.; Piotrowska, A.; Frydlewicz, A.; Karczewski, M.; Dziegiel, P.; Witkiewicz, W. Correlation of HIWI and HILI Expression with Cancer Stem Cell Markers in Colorectal Cancer. Anticancer Res. 2015, 35, 3317–3324. [Google Scholar]
- Hutter, C.; Zenklusen, J.C. The Cancer Genome Atlas: Creating Lasting Value beyond Its Data. Cell 2018, 173, 283–285. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Barault, L.; Amatu, A.; Siravegna, G.; Ponzetti, A.; Moran, S.; Cassingena, A.; Mussolin, B.; Falcomata, C.; Binder, A.M.; Cristiano, C.; et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut 2018, 67, 1995–2005. [Google Scholar] [CrossRef] [PubMed]
- Shaiken, T.E.; Opekun, A.R. Dissecting the cell to nucleus, perinucleus and cytosol. Sci. Rep. 2014, 4, 4923. [Google Scholar] [CrossRef] [Green Version]
- Locati, M.D.; Terpstra, I.; De Leeuw, W.C.; Kuzak, M.; Rauwerda, H.; Ensink, W.A.; Van Leeuwen, S.; Nehrdich, U.; Spaink, H.P.; Jonker, M.J.; et al. Improving small RNA-seq by using a synthetic spike-in set for size-range quality control together with a set for data normalization. Nucleic Acids Res. 2015, 43, e89. [Google Scholar] [CrossRef]
- Keene, J.D.; Komisarow, J.M.; Friedersdorf, M.B. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006, 1, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Ko, E.-A.; Sanders, K.M.; Chen, Q.; Zhou, T. SPORTS1.0: A Tool for Annotating and Profiling Non-coding RNAs Optimized for rRNA- and tRNA-derived Small RNAs. Genom. Proteom. Bioinform. 2018, 16, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Panero, R.; Rinaldi, A.; Memoli, D.; Nassa, G.; Ravo, M.; Rizzo, F.; Tarallo, R.; Milanesi, L.; Weisz, A.; Giurato, G. iSmaRT: A toolkit for a comprehensive analysis of small RNA-Seq data. Bioinformatics 2016, 33, 938–940. [Google Scholar]
- Yagi, K.; Akagi, K.; Hayashi, H.; Nagae, G.; Tsuji, S.; Isagawa, T.; Midorikawa, Y.; Nishimura, Y.; Sakamoto, H.; Seto, Y.; et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin. Cancer Res. 2010, 16, 21–33. [Google Scholar] [CrossRef]
- Navarro, A.; Tejero, R.; Viñolas, N.; Cordeiro, A.; Marrades, R.M.; Fuster, D.; Caritg, O.; Moises, J.; Muñoz, C.; Molins, L.; et al. The significance of PIWI family expression in human lung embryogenesis and non-small cell lung cancer. Oncotarget 2015, 6, 31544–31556. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, H.J.; Heyn, H.; Garcia del Muro, X.; Vidal, A.; Larriba, S.; Munoz, C.; Villanueva, A.; Esteller, M. Epigenetic loss of the PIWI/piRNA machinery in human testicular tumorigenesis. Epigenetics 2014, 9, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gu, Y.; Zhang, K.; Xie, K.; Zhu, M.; Dai, N.; Jiang, Y.; Guo, X.; Liu, M.; Dai, J.; et al. Systematic identification of genes with a cancer-testis expression pattern in 19 cancer types. Nat. Commun. 2016, 7, 10499. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Zhang, K.; Kong, J.; Wang, C.; Gu, Y.; Liang, C.; Jiang, T.; Qin, N.; Liu, J.; Guo, X.; et al. Cancer-testis gene PIWIL1 promotes cell proliferation, migration, and invasion in lung adenocarcinoma. Cancer Med. 2018, 7, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chiappinelli, K.B.; Guzzetta, A.A.; Easwaran, H.; Yen, R.-W.C.; Vatapalli, R.; Topper, M.J.; Luo, J.; Connolly, R.M.; Azad, N.S.; et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 2014, 5, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef]
- Lappalainen, I.; Almeida-King, J.; Kumanduri, V.; Senf, A.; Spalding, J.D.; Ur-Rehman, S.; Saunders, G.; Kandasamy, J.; Caccamo, M.; Leinonen, R.; et al. The European Genome-phenome Archive of human data consented for biomedical research. Nat. Genet. 2015, 47, 692–695. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Wang, J.; Xu, J.; Zhang, Z.; Koppetsch, B.S.; Schultz, N.; Vreven, T.; Meignin, C.; Davis, I.; Zamore, P.D.; et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 2012, 151, 871–884. [Google Scholar] [CrossRef]
- Genzor, P.; Cordts, S.C.; Bokil, N.V.; Haase, A.D. Aberrant expression of select piRNA-pathway genes does not reactivate piRNA silencing in cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 11111–11112. [Google Scholar] [CrossRef] [Green Version]
- Vagin, V.V.; Wohlschlegel, J.; Qu, J.; Jónsson, Z.; Huang, X.; Chuma, S.; Girard, A.; Sachidanandam, R.; Hannon, G.J.; Aravin, A.A. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genome Res. 2009, 23, 1749–1762. [Google Scholar] [CrossRef] [Green Version]
- Ebhardt, H.A.; Thi, E.P.; Wang, M.-B.; Unrau, P.J. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl. Acad. Sci. USA 2005, 102, 13398–13403. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, D.G.; Teysset, L.; Carré, C. RNA 2′-O-Methylation (Nm) Modification in Human Diseases. Genes 2019, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- De Fazio, S.; Bartonicek§, N.; Di Giacomo, M.; Abreu-Goodger, C.; Sankar, A.; Funaya, C.; Antony, C.; Moreira, P.N.; Enright, A.J.; O’Carroll, D. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 2011, 480, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Gomes Fernandes, M.; He, N.; Wang, F.; Van Iperen, L.; Eguizabal, C.; Matorras, R.; Roelen, B.A.J.; Chuva De Sousa Lopes, S.M. Human-specific subcellular compartmentalization of P-element induced wimpy testis-like (PIWIL) granules during germ cell development and spermatogenesis. Hum. Reprod. 2018, 33, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.; Zhou, N.; Wu, K.; Guo, Y.; Tan, W.; Zhang, H.; Zhang, X.; Geng, G.; Pan, T.; Luo, H.; et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015, 43, 10474–10491. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Chen, X.; Zhang, X.; Duan, X.; Pan, T.; Hu, Q.; Zhang, Y.; Zhong, F.; Liu, J.; Zhang, H.; et al. An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. 2015, 43, 3712–3725. [Google Scholar] [CrossRef] [PubMed]
- Franz, M.; Rodríguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef] [Green Version]
- Weng, W.; Li, H.; Goel, A. Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications. Biochim. et Biophys. Acta (BBA) Bioenerg. 2019, 1871, 160–169. [Google Scholar] [CrossRef]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef]
- Kabayama, Y.; Toh, H.; Katanaya, A.; Sakurai, T.; Chuma, S.; Kuramochi-Miyagawa, S.; Saga, Y.; Nakano, T.; Sasaki, H. Roles of MIWI, MILI and PLD6 in small RNA regulation in mouse growing oocytes. Nucleic Acids Res. 2017, 45, 5387–5398. [Google Scholar] [CrossRef] [Green Version]
- Viljetic, B.; Diao, L.; Liu, J.; Krsnik, Z.; Wijeratne, S.H.R.; Kristopovich, R.; Dutre-Clarke, M.; Kraushar, M.L.; Song, J.; Xing, J.; et al. Multiple roles of PIWIL1 in mouse neocorticogenesis. bioRxiv 2017, 106070. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, F.; Hashim, A.; Marchese, G.; Ravo, M.; Tarallo, R.; Nassa, G.; Giurato, G.; Rinaldi, A.; Cordella, A.; Persico, M.; et al. Timed regulation of P-element-induced wimpy testis-interacting RNA expression during rat liver regeneration. Hepatology 2014, 60, 798–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamore, P.D. Somatic piRNA biogenesis. EMBO J. 2010, 29, 3219–3221. [Google Scholar] [CrossRef] [Green Version]
- Théron, E.; Dennis, C.; Brasset, E.; Vaury, C. Distinct features of the piRNA pathway in somatic and germ cells: From piRNA cluster transcription to piRNA processing and amplification. Mob. DNA 2014, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, C.; Theurkauf, W. Biogenesis and germline functions of piRNAs. Development 2008, 135, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Klattenhoff, C.; Xi, H.; Li, C.; Lee, S.; Xu, J.; Khurana, J.S.; Zhang, F.; Schultz, N.; Koppetsch, B.S.; Nowosielska, A.; et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 2009, 138, 1137–1149. [Google Scholar] [CrossRef]
- Yin, J.; Qi, W.; Ji, C.; Zhang, D.; Xie, X.; Ding, Q.; Jiang, X.; Han, J.; Jiang, H. Small RNA sequencing revealed aberrant piRNA expression profiles in colorectal cancer. Oncol. Rep. 2019, 42, 263–272. [Google Scholar] [CrossRef]
- Gou, L.-T.; Dai, P.; Yang, J.-H.; Xue, Y.; Hu, Y.-P.; Zhou, Y.; Kang, J.-Y.; Wang, X.; Li, H.; Hua, M.-M.; et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014, 24, 680–700. [Google Scholar] [CrossRef]
- Vourekas, A.; Zheng, Q.; Alexiou, P.; Maragkakis, M.; Kirino, Y.; Gregory, B.D.; Mourelatos, Z. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat. Struct. Mol. Boil. 2012, 19, 773–781. [Google Scholar] [CrossRef] [Green Version]
- Popovici, V.; Budinska, E.; Tejpar, S.; Weinrich, S.; Estrella, H.; Hodgson, G.; Van Cutsem, E.; Xie, T.; Bosman, F.T.; Roth, A.D.; et al. Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J. Clin. Oncol. 2012, 30, 1288–1295. [Google Scholar] [CrossRef]
- Herr, R.; Kohler, M.; Andrlova, H.; Weinberg, F.; Moller, Y.; Halbach, S.; Lutz, L.; Mastroianni, J.; Klose, M.; Bittermann, N.; et al. B-Raf inhibitors induce epithelial differentiation in BRAF-mutant colorectal cancer cells. Cancer Res. 2015, 75, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Bi, L.; Liu, Q.; Zhao, M.; Cao, B.; Li, D.; Xiu, J. Hiwi Promotes the Proliferation of Colorectal Cancer Cells via Upregulating Global DNA Methylation. Dis. Markers 2015, 2015, 383056. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, S.; Terry, M.; Matushansky, I. Hiwi mediated tumorigenesis is associated with DNA hypermethylation. PLoS ONE 2012, 7, e33711. [Google Scholar] [CrossRef] [PubMed]
- Piskol, R.; Huw, L.Y.; Sergin, I.; Kljin, C.; Modrusan, Z.; Kim, D.; Kljavin, N.M.; Tam, R.; Patel, R.; Burton, J.; et al. A Clinically Applicable Gene-Expression Classifier Reveals Intrinsic and Extrinsic Contributions to Consensus Molecular Subtypes in Primary and Metastatic Colon Cancer. Clin. Cancer Res. 2019, 25, 4431–4442. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| PIWIL1 | ACGAAGTGCCACAGTTTTTGG | AGTCTTCCTCCAGACTGAGC |
| PIWIL2 | GCCTGGGTTGAACTAAAGGA | CCATGATGATGCAAACAACC |
| PIWIL3 | TCAGATGGCAGCAAAATCAC | ACGTTGTGTACCCGTTAGGC |
| PIWIL4 | ATGGCACCGAGATCACCTAT | GCTGAGCCTCACTGTTGTCA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sellitto, A.; Geles, K.; D’Agostino, Y.; Conte, M.; Alexandrova, E.; Rocco, D.; Nassa, G.; Giurato, G.; Tarallo, R.; Weisz, A.; et al. Molecular and Functional Characterization of the Somatic PIWIL1/piRNA Pathway in Colorectal Cancer Cells. Cells 2019, 8, 1390. https://doi.org/10.3390/cells8111390
Sellitto A, Geles K, D’Agostino Y, Conte M, Alexandrova E, Rocco D, Nassa G, Giurato G, Tarallo R, Weisz A, et al. Molecular and Functional Characterization of the Somatic PIWIL1/piRNA Pathway in Colorectal Cancer Cells. Cells. 2019; 8(11):1390. https://doi.org/10.3390/cells8111390
Chicago/Turabian StyleSellitto, Assunta, Konstantinos Geles, Ylenia D’Agostino, Marisa Conte, Elena Alexandrova, Domenico Rocco, Giovanni Nassa, Giorgio Giurato, Roberta Tarallo, Alessandro Weisz, and et al. 2019. "Molecular and Functional Characterization of the Somatic PIWIL1/piRNA Pathway in Colorectal Cancer Cells" Cells 8, no. 11: 1390. https://doi.org/10.3390/cells8111390
APA StyleSellitto, A., Geles, K., D’Agostino, Y., Conte, M., Alexandrova, E., Rocco, D., Nassa, G., Giurato, G., Tarallo, R., Weisz, A., & Rizzo, F. (2019). Molecular and Functional Characterization of the Somatic PIWIL1/piRNA Pathway in Colorectal Cancer Cells. Cells, 8(11), 1390. https://doi.org/10.3390/cells8111390