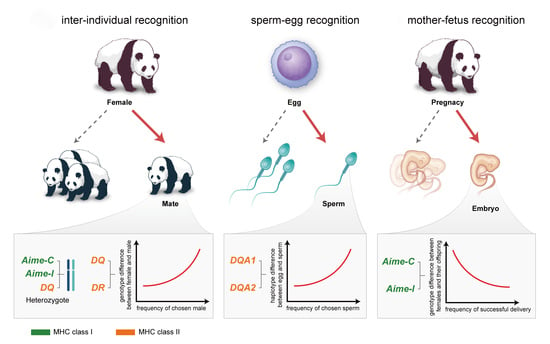
Reproductive Strategy Inferred from Major Histocompatibility Complex-Based Inter-Individual, Sperm-Egg, and Mother-Fetus Recognitions in Giant Pandas (Ailuropoda melanoleuca)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Species and Behavioral Observations of Inter-Individual Recognition
2.2. Sperm-Egg and Mother-Fetus Recognition
2.3. DNA Extraction, MHC and Microsatellite Genotyping, and the Definition of a Super Haplotype
2.4. Paternity Test
2.5. Data Analysis
2.5.1. Female Mate Choice at the Inter-Individual Recognition Level
2.5.2. Sperm-Egg Recognition Level
2.5.3. Mother-Fetus Recognition Level
2.6. Declarations Ethics Statement
3. Results
3.1. Microsatellite and MHC Diversity
3.2. Inter-Individual Recognition
3.2.1. Heterozygosity Advantage
Males Involved in Natural Mating Versus Randomly Assigned Males
Males Involved in Natural Mating Versus Natural Non-Mating Males
Relationship between Male MHC Heterozygosity and Natural Mating Success
3.2.2. Genetic Compatibility and Inbreeding Avoidance
Naturally Mated Pairs versus Randomly Assigned Pairs
Naturally Mated Pairs versus Non-Mating Pairs
Relationship between MHC Divergence between Females and Males and Natural Mating Success
3.3. Sperm-Egg Recognition
3.3.1. Observed Zygotes (Offspring) Versus Randomly Assigned Zygotes
3.3.2. Observed Zygotes (Offspring) Versus Other Zygotes
3.3.3. Relationship between Breeding Success and MHC Divergence of Zygotes
3.4. Mother-Fetus Recognition
4. Discussion
4.1. Female Choice at the Inter-Individual Recognition Level
4.2. Cryptic Female Choice at the Sperm-Egg Recognition Level
4.3. Mother-Fetus Recognition Level
4.4. Hierarchical and Cooperative Effects
4.5. Relative Importance of MHC-I, MHC-II, DQ, and DR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Andersson, M.B. Sexual Selection; Princeton University Press: Princeton, NJ, USA, 1994. [Google Scholar]
- Andersson, M.; Simmons, L.W. Sexual selection and mate choice. Trends Ecol. Evol. 2006, 21, 296–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jennions, M.D.; Petrie, M. Why do females mate multiply? A review of the genetic benefits. Biol. Rev. 2000, 75, 21–64. [Google Scholar] [CrossRef]
- Searcy, W.A. The evolutionary effects of mate selection. Annu. Rev. Ecol. Syst. 1982, 13, 57–85. [Google Scholar] [CrossRef]
- Folstad, I.; Karter, A.J. Parasites, bright males, and the immunocompetence handicap. Am. Nat. 1992, 603–622. [Google Scholar] [CrossRef]
- Hamilton, W.D.; Zuk, M. Heritable true fitness and bright birds: A role for parasites? Science 1982, 218, 384–387. [Google Scholar] [CrossRef]
- Klein, J. The Natural History of the Major Histocompatibility Complex; Wiley & Sons: New York, NY, USA, 1986. [Google Scholar]
- Sommer, S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front. Zool. 2005, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Piertney, S.B.; Oliver, M.K. The evolutionary ecology of the major histocompatibility complex. Heredity 2006, 96, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Milinski, M. The major histocompatibility complex, sexual selection, and mate choice. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 159–186. [Google Scholar] [CrossRef]
- de Campos-Lima, P.O.; Gavioli, R.; Zhang, Q.J.; Wallace, L.E.; Dolcetti, R.; Rowe, M.; Rickinson, A.B.; Masucci, M.G. HLA-A11 epitope loss isolates of Epstein-Barr virus from a highly A11+ population. Science 1993, 260, 98–100. [Google Scholar] [CrossRef] [PubMed]
- Grimholt, U.; Larsen, S.; Nordmo, R.; Midtlyng, P.; Kjoeglum, S.; Storset, A.; Saebø, S.; Stet, R.J.M. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics 2003, 55, 210–219. [Google Scholar] [CrossRef]
- Harf, R.; Sommer, S. Association between major histocompatibility complex class II DRB alleles and parasite load in the hairy-footed gerbil, Gerbillurus paeba, in the southern Kalahari. Mol. Ecol. 2004, 14, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Langefors, Å.; Lohm, J.; Grahn, M.; Andersen, Ø.; von Schantz, T. Association between major histocompatibility complex class IIB alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proc. R. Soc. Lond. B Biol. Sci. 2001, 268, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Mays, H.L.; Hill, G.E. Choosing mates: Good genes versus genes that are a good fit. Trends Ecol. Evol. 2004, 19, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Neff, B.D.; Pitcher, T.E. Genetic quality and sexual selection: An integrated framework for good genes and compatible genes. Mol. Ecol. 2005, 14, 19–38. [Google Scholar] [CrossRef] [PubMed]
- Cutrera, A.P.; Fanjul, M.S.; Zenuto, R.R. Females prefer good genes: MHC-associated mate choice in wild and captive tuco-tucos. Anim. Behav. 2012, 83, 847–856. [Google Scholar] [CrossRef]
- Promerová, M.; Vinkler, M.; Bryja, J.; Poláková, R.; Schnitzer, J.; Munclinger, P.; Albrecht, T. Occurrence of extra-pair paternity is connected to social male’s MHC-variability in the scarlet rosefinch Carpodacus erythrinus. J. Avian Biol. 2011, 42, 5–10. [Google Scholar] [CrossRef]
- Tregenza, T.; Wedell, N. Genetic compatibility, mate choice and patterns of parentage: Invited review. Mol. Ecol. 2000, 9, 1013–1027. [Google Scholar] [CrossRef] [PubMed]
- Schwensow, N.; Eberle, M.; Sommer, S. Compatibility counts: MHC-associated mate choice in a wild promiscuous primate. Proc. R. Soc. B Biol. Sci. 2008, 275, 555–564. [Google Scholar] [CrossRef]
- Strandh, M.; Westerdahl, H.; Pontarp, M.; Canback, B.; Dubois, M.P.; Miquel, C.; Taberlet, P.; Bonadonna, F. Major histocompatibility complex class II compatibility, but not class I, predicts mate choice in a bird with highly developed olfaction. Proc. R. Soc. B Biol. Sci. 2012, 279, 4457–4463. [Google Scholar] [CrossRef]
- Juola, F.A.; Dearborn, D.C. Sequence-based evidence for major histocompatibility complex-disassortative mating in a colonial seabird. Proc. R. Soc. B Biol. Sci. 2012, 279, 153–162. [Google Scholar] [CrossRef]
- Reichard, M.; Spence, R.; Bryjova, A.; Bryja, J.; Smith, C. Female Rose Bitterling Prefer MHC-Dissimilar Males: Experimental Evidence. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Bahr, A.; Sommer, S.; Mattle, B.; Wilson, A.B. Mutual mate choice in the potbellied seahorse (Hippocampus abdominalis). Behav. Ecol. 2012, 23, 869–878. [Google Scholar] [CrossRef]
- Schwensow, N.; Fietz, J.; Dausmann, K.; Sommer, S. MHC-associated mating strategies and the importance of overall genetic diversity in an obligate pair-living primate. Evol. Ecol. 2008, 22, 617–636. [Google Scholar] [CrossRef]
- Setchell, J.; Charpentier, M.; Abbott, K.; Wickings, E.; Knapp, L. Opposites attract: MHC-associated mate choice in a polygynous primate. J. Evol. Biol. 2010, 23, 136–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reusch, T.B.H.; Haberli, M.; Aeschlimann, P.B.; Milinski, M. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature 2001, 414, 300–302. [Google Scholar] [CrossRef] [PubMed]
- Aeschlimann, P.B.; Haberli, M.A.; Reusch, T.B.H.; Boehm, T.; Milinski, M. Female sticklebacks Gasterosteus aculeatus use self-reference to optimize MHC allele number during mate selection. Behav. Ecol. Sociobiol. 2003, 54, 119–126. [Google Scholar] [CrossRef]
- Arkush, K.D.; Giese, A.R.; Mendonca, H.L.; McBride, A.M.; Marty, G.D.; Hedrick, P.W. Resistance to three pathogens in the endangered winter-run chinook salmon (Oncorhynchus tshawytscha): Effects of inbreeding and major histocompatibility complex genotypes. Can. J. Fish. Aquat. Sci. 2002, 59, 966–975. [Google Scholar] [CrossRef]
- Meagher, S.; Penn, D.J.; Potts, W.K. Male-Male Competition Magnifies Inbreeding Depression in Wild House Mice. Proc. Natl. Acad. Sci. USA 2000, 97, 3324–3329. [Google Scholar] [CrossRef]
- MartinVilla, J.M.; Luque, I.; MartinezQuiles, N.; Corell, A.; Regueiro, J.R.; Timon, M.; ArnaizVillena, A. Diploid expression of human leukocyte antigen class I and class II molecules on spermatozoa and their cyclic inverse correlation with inhibin concentration. Biol. Reprod. 1996, 55, 620–629. [Google Scholar] [CrossRef] [Green Version]
- Martin-Villa, J.M.; Longas, J.; Arnaiz-Villena, A. Cyclic expression of HLA class I and II molecules on the surface of purified human spermatozoa and their control by serum inhibin B levels. Biol. Reprod. 1999, 61, 1381–1386. [Google Scholar] [CrossRef]
- Paradisi, R.; Neri, S.; Pession, A.; Magrini, E.; Bellavia, E.; Ceccardi, S.; Flamigni, C. Human leukocyte antigen II expression in sperm cells: Comparison between fertile and infertile men. Arch. Androl. 2000, 45, 203–213. [Google Scholar] [PubMed]
- Rulicke, T.; Chapuisat, M.; Homberger, F.R.; Macas, E.; Wedekind, C. MHC-genotype of progeny influenced by parental infection. Proc. R. Soc. B Biol. Sci. 1998, 265, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Knapp, L.A.; Ha, J.C.; Sackett, G.P. Parental MHC antigen sharing and pregnancy wastage in captive pigtailed macaques. J. Reprod. Immunol. 1996, 32, 73–88. [Google Scholar] [CrossRef]
- Ober, C.; Weitkamp, L.R.; Cox, N.; Dytch, H.; Kostyu, D.; Elias, S. HLA and mate choice in humans. Am. J. Hum. Genet. 1997, 61, 497–504. [Google Scholar] [CrossRef]
- Schacter, B.; Weitkamp, L.R.; Johnson, W.E. Parental HLA compatibility, fetal wastage and neural-tube defects-evidence for a T/T-linke locus in humans. Am. J. Hum. Genet. 1984, 36, 1082–1091. [Google Scholar]
- Lovlie, H.; Gillingham, M.A.F.; Worley, K.; Pizzari, T.; Richardson, D.S. Cryptic female choice favours sperm from major histocompatibility complex-dissimilar males. Proc. R. Soc. B Biol. Sci. 2013, 280, 20131296. [Google Scholar] [CrossRef]
- Yeates, S.; Einum, S.; Fleming, I.; Megens, H.-J.; Hindar, K.; Holt, W.; Van Look, K.; Gage, M. Atlantic salmon eggs favour sperm in competition that have similar major histocompatibility alleles. Proc. R. Soc. Lond. B Biol. Sci. 2009, 276, 559–566. [Google Scholar] [CrossRef]
- Gessner, C.; Nakagawa, S.; Zavodna, M.; Gemmell, N.J. Sexual selection for genetic compatibility: The role of the major histocompatibility complex on cryptic female choice in Chinook salmon (Oncorhynchus tshawytscha). Heredity 2017, 118. [Google Scholar] [CrossRef]
- Gasparini, C.; Congiu, L.; Pilastro, A. Major histocompatibility complex similarity and sexual selection: Different does not always mean attractive. Mol. Ecol. 2015, 24, 4286–4295. [Google Scholar] [CrossRef]
- Lenz, T.L.; Hafer, N.; Samonte, I.E.; Yeates, S.E.; Milinski, M. Cryptic haplotype-specific gamete selection yields offspring with optimal MHC immune genes. Evolution 2018, 72, 2478–2490. [Google Scholar] [CrossRef]
- Hu, J.C. Research on the Giant Panda; Shanghai Publishing House of Science and Technology: Shanghai, China, 2001. [Google Scholar]
- Martin-Wintle, M.S.; Shepherdson, D.; Zhang, G.; Zhang, H.; Li, D.; Zhou, X.; Li, R.; Swaisgood, R.R. Free mate choice enhances conservation breeding in the endangered giant panda. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Wang, P.Y. The Study on Reproduction in Giant Panda; China FORESTRY publishing House: Bei Jing, China, 2003. [Google Scholar]
- Feng, W.H.; Zhang, A.J. The Research on Breeding and Disease of Giant Panda; Sichuan Publishing House of Science and Technology: Cheng Du, China, 1991. [Google Scholar]
- Huchard, E.; Baniel, A.; Schliehe-Diecks, S.; Kappeler, P.M. MHC-disassortative mate choice and inbreeding avoidance in a solitary primate. Mol. Ecol. 2013, 22, 4071–4086. [Google Scholar] [CrossRef] [PubMed]
- Babik, W. Methods for MHC genotyping in non-model vertebrates. Mol. Ecol. Resour. 2010, 10, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Nie, Y.; Yan, L.; Hu, Y.; Wei, F. No evidence for MHC-based mate choice in wild giant pandas. Ecol. Evol. 2018, 8, 8642–8651. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.H.; Zeng, C.J.; Ni, X.W.; Pan, H.J.; Fang, S.G. Giant panda genomic data provide insight into the birth-and-death process of mammalian major histocompatibility complex class II genes. PLoS ONE 2009, 4, e4147. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.H.; Zhang, P.; Ni, X.W.; Wu, H.L.; Chen, Y.Y.; Kuang, Y.Y.; Ge, Y.F.; Fang, S.G. A novel HURRAH protocol reveals high numbers of monomorphic MHC class II loci and two asymmetric multi-locus haplotypes in the Père David’s deer. PLoS ONE 2011, 6, e14518. [Google Scholar] [CrossRef]
- Zhu, Y.; Sun, D.D.; Ge, Y.F.; Yu, B.; Chen, Y.Y.; Wan, Q.H. Isolation and characterization of class I MHC genes in the giant panda (Ailuropoda melanoleuca). Chin. Sci. Bull. 2012, 57, 1–8. [Google Scholar] [CrossRef]
- Xie, Z.; Gipps, J. The 2009 International Studbood for Giant Panda (Ailuropoda melanoleuca); Chinese Association of Zoological Garden: Beijing, China, 2009. [Google Scholar]
- Durrant, B.S.; Olson, M.A.; Amodeo, D.; Anderson, A.; Russ, K.D.; Campos-Morales, R.; Gual-Sill, F.; Garza, J.R. Vaginal cytology and vulvar swelling as indicators of impending estrus and ovulation in the giant panda (Ailuropoda melanoleuca). Zoo Biol. 2003, 22, 313–321. [Google Scholar] [CrossRef]
- Owen, M.A.; Swaisgood, R.R.; McGeehan, L.; Zhou, X.P.; Lindburg, D.G. Dynamics of Male-Female Multimodal Signaling Behavior across the Estrous Cycle in Giant Pandas (Ailuropoda melanoleuca). Ethology 2013, 119, 869–880. [Google Scholar] [CrossRef]
- Wan, Q.H.; Zhu, L.; Wu, H.; Fang, S.G. Major histocompatibility complex class II variation in the giant panda (Ailuropoda melanoleuca). Mol. Ecol. 2006, 15, 2441–2450. [Google Scholar] [CrossRef]
- Li, D.; Cui, H.; Wang, C.; Ling, S.; Huang, Z.; Zhang, H. A fast and effective method to perform paternity testing for Wolong giant pandas. Chin. Sci. Bull. 2011, 56, 2559–2564. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.M.; Guo, Y.; Li, D.S.; Wang, P.Y.; Fang, S.G. Sixteen novel microsatellite loci developed for the giant panda (Ailuropoda melanoleuca). Conserv. Genet. 2009, 10, 589–592. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Zhu, Y.; Wan, Q.H.; Lou, J.K.; Li, W.J.; Ge, Y.F.; Fang, S.G. Patterns of adaptive and neutral diversity identify the Xiaoxiangling Mountains as a refuge for the giant panda. PLoS ONE 2013. [Google Scholar] [CrossRef]
- Zeng, C.J.; Pan, H.J.; Gong, S.B.; Yu, J.Q.; Wan, Q.H.; Fang, S.G. Giant panda BAC library construction and assembly of a 650-kb contig spanning major histocompatibility complex class II region. BMC Genom. 2007, 8, 315. [Google Scholar] [CrossRef]
- Coltman, D.; Slate, J. Microsatellite measures of inbreeding: A meta-analysis. Evolution 2003, 57, 971–983. [Google Scholar] [CrossRef]
- Wetton, J.H.; Carter, R.E.; Parkin, D.T.; Walters, D. Demographic study of a wild house sparrow population by DNA fingerprinting. Nature 1987, 327, 147–149. [Google Scholar] [CrossRef]
- Landry, C.; Garant, D.; Duchesne, P.; Bernatchez, L. ‘Good genes as heterozygosity’: The major histocompatibility complex and mate choice in Atlantic salmon (Salmo salar). Proc. R. Soc. Lond. B Biol. Sci. 2001, 268, 1279–1285. [Google Scholar] [CrossRef]
- Sandberg, M.; Eriksson, L.; Jonsson, J.; Sjöström, M.; Wold, S. New chemical descriptors relevant for the design of biologically active peptides. A multivariate characterization of 87 amino acids. J. Med. Chem. 1998, 41, 2481–2491. [Google Scholar] [CrossRef]
- Huchard, E.; Weill, M.; Cowlishaw, G.; Raymond, M.; Knapp, L.A. Polymorphism, haplotype composition, and selection in the Mhc-DRB of wild baboons. Immunogenetics 2008, 60, 585–598. [Google Scholar] [CrossRef]
- Hill, G.E. Plumage coloration is a sexually selected indicator of male quality. Nature 1991, 350, 337–339. [Google Scholar] [CrossRef]
- Kurtz, J.; Wegner, K.M.; Kalbe, M.; Reusch, T.B.; Schaschl, H.; Hasselquist, D.; Milinski, M. MHC genes and oxidative stress in sticklebacks: An immuno-ecological approach. Proc. Rroc. Soc. B Biol. Sci. 2006, 273, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Bjorkman, P.J.; Saper, M.A.; Samraoui, B.; Bennett, W.S.; Strominger, J.L.; Wiley, D.C. The foreigh antigen bing site and T cell recognition regions of class I histocompatibility antigens. Nat. Immunol. 1987, 329, 512–518. [Google Scholar]
- Queller, D.C.; Goodnight, K.F. Estimating relatedness using genetic markers. Evolution 1989, 258–275. [Google Scholar] [CrossRef] [PubMed]
- Hardy, O.J.; Vekemans, X. SPAGeDi: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yekutieli, D.; Benjamini, Y. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J. Stat. Plan. Infer. 1999, 82, 171–196. [Google Scholar] [CrossRef]
- McGraw, K.J.; Hill, G.E. Differential effects of endoparasitism on the expression of carotenoid-and melanin-based ornamental coloration. Proc. R. Soc. Lond. B Biol. Sci. 2000, 267, 1525–1531. [Google Scholar] [CrossRef] [PubMed]
- Fitze, P.S.; Richner, H. Differential effects of a parasite on ornamental structures based on melanins and carotenoids. Behav. Ecol. 2002, 13, 401–407. [Google Scholar] [CrossRef]
- Siefferman, L.; Hill, G.E. Structural and melanin coloration indicate parental effort and reproductive success in male eastern bluebirds. Behav. Ecol. 2003, 14, 855–861. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.; Magurran, A. Multiple benefits of multiple mating in guppies. Proc. Natl. Acad. Sci. USA 2000, 97, 10074–10076. [Google Scholar] [CrossRef] [Green Version]
- Colegrave, N.; Kotiaho, J.S.; Tomkins, J.L. Mate choice or polyandry: Reconciling genetic compatibility and good genes sexual selection. Evol. Ecol. Res. 2002, 4, 911–917. [Google Scholar]
- Roberts, S.C.; Gosling, L.M. Genetic similarity and quality interact in mate choice decisions by female mice. Nat. Genet. 2003, 35, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Lopez, P.; Martin, J. Pheromonal recognition of females takes precedence over the chromatic cue in male Iberian wall lizards Podarcis hispanica. Ethology 2001, 107, 901–912. [Google Scholar] [CrossRef]
- Candolin, U. The use of multiple cues in mate choice. Biol. Rev. 2003, 78, 575–595. [Google Scholar] [CrossRef] [Green Version]
- Huchard, E.; Knapp, L.A.; Wang, J.; Raymond, M.; Cowlishaw, G. MHC, mate choice and heterozygote advantage in a wild social primate. Mol. Ecol. 2010, 19, 2545–2561. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.H.; Wang, R.L.; Zhong, S.M.; Ye, Z.Y.; Cui, X.Z.; Zeng, J.H. Analysis on the Dead Cause of the Anatomical Carcass of Giant Panda (Ailuropoda Melanoleuca); Sichuan Scientific & Technical Publishers: Chengdu, China, 1991; pp. 244–248. [Google Scholar]
- Ye, Z.Y. The Control of the Diseases of Giant Panda in Field: Report of 50 Cases; Sichuan Scientific & Technical Publishers: Chengdu, China, 1991. [Google Scholar]
- Mainka, S.A.; Qiu, X.M.; He, T.M.; Appel, M.J. Serologic survey of giant pandas (Ailuropoda melanoleuca), and domestic dogs and cats in the Wolong Reserve, China. J. Wildl. Dis. 1994, 30, 86–89. [Google Scholar] [CrossRef]
- Qin, Q.; Li, D.S.; Zhang, H.M.; Hou, R.; Zhang, Z.H.; Zhang, C.L.; Zhang, J.G.; Wei, F.W. Serosurvey of selected viruses in captive giant pandas ( Ailuropoda melanoleuca) in China. Vet. Microbiol. 2010, 142, 199–204. [Google Scholar] [CrossRef]
- Fedorka, K.M.; Mousseau, T.A. Material and genetic benefits of female multiple mating and polyandry. Anim. Behav. 2002, 64, 361–367. [Google Scholar] [CrossRef]
- Yasui, Y. A" good-sperm" model can explain the evolution of costly multiple mating by females. Am. Nat. 1997, 149, 573–584. [Google Scholar] [CrossRef]
- Schmidt, C.M.; Orr, H.T. Maternal/fetal interactions: The role of the MHC class I molecule HLA-G. Crit. Rev. Immunol. 1992, 13, 207–224. [Google Scholar]
- Rouas-Freiss, N.; GoncAlves, R.M.-B.; Menier, C.; Dausset, J.; Carosella, E.D. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc. Natl. Acad. Sci. USA 1997, 94, 11520–11525. [Google Scholar] [CrossRef] [PubMed] [Green Version]









| Compatibility Test | Heterozygote Advantage Test | |||
|---|---|---|---|---|
| Relatedness | MLH | d2 | ||
| Randomization | Observed | −0.087 | 0.751 | 25.961 |
| Test a | Simulated mean | −0.065 | 0.744 | 27.491 |
| 95% CI | [−0.090, −0.040] | [0.713, 0.775] | [24.319, 30.648] | |
| P | 0.079 | 0.651 | 0.345 | |
| Paired test b | Potential fathers | −0.093 | 0.782 | 25.157 |
| Other males | −0.061 | 0.741 | 27.480 | |
| Statistics | t = 1.592 | Z = 0.556 | t = 0.913 | |
| P | 0.115 | 0.578 | 0.364 | |
| Locus | Region | Simulated Mean [95% CI] | Observe Mean | P |
|---|---|---|---|---|
| SuHa | ABS | 123.873 [121.759, 125.899] | 127.832 | 0.001 |
| ALL | 136.412 [134.088, 138.677] | 140.728 | 0.000 | |
| SuHaI | ABS | 93.169 [90.939, 95.359] | 94.326 | 0.297 |
| ALL | 101.310 [98.883, 103.635] | 102.285 | 0.416 | |
| SuHaII | ABS | 73.374 [71.216, 75.579] | 78.033 | 0.000 |
| ALL | 82.600 [80.143, 85.028] | 87.462 | 0.000 | |
| DQ | ABS | 53.815 [51.715, 55.876] | 58.000 | 0.000 |
| ALL | 57.319 [55.059, 59.577] | 61.433 | 0.001 | |
| DR | ABS | 45.051 [43.434, 46.662] | 47.498 | 0.002 |
| ALL | 53.531 [51.588, 55.516] | 56.510 | 0.003 | |
| C | ABS | 60.340 [58.005, 62.674] | 62.840 | 0.036 |
| ALL | 63.839 [61.359, 66.265] | 66.431 | 0.038 | |
| I | ABS | 46.787 [44.406, 49.218] | 45.218 | 0.196 |
| ALL | 50.388 [47.956, 52.819] | 48.681 | 0.172 | |
| L | ABS | 33.315 [31.085, 35.588] | 32.624 | 0.548 |
| ALL | 39.862 [37.267, 42.449] | 38.584 | 0.340 | |
| DQA1 | ABS | 32.307 [30.740, 33.938] | 35.587 | 0.000 |
| ALL | 35.121 [30.740,33.938] | 38.400 | 0.000 | |
| DQA2 | ABS | 11.596 [10.674, 12.542] | 13.076 | 0.001 |
| ALL | 11.603 [10.674, 12.542] | 13.076 | 0.001 | |
| DQB1 | ABS | 32.821 [30.624, 35.005] | 36.002 | 0.005 |
| ALL | 34.555 [32.231, 36.817] | 37.924 | 0.003 | |
| DRB3 | ABS | 45.051 [43.434, 46.662] | 47.498 | 0.002 |
| ALL | 53.531 [51.588, 55.516] | 56.510 | 0.003 |
| Locus | Natural Mated Pairs | Non-Mating Pairs | Statistics | P | |
|---|---|---|---|---|---|
| SuHa | ABS | 126.209 | 119.677 | t = 3.310 | 0.001 |
| ALL | 138.990 | 132.018 | t = 3.298 | 0.001 | |
| SuHaI | ABS | 92.315 | 90.284 | t = 1.054 | 0.294 |
| ALL | 100.215 | 98.262 | t = 0.932 | 0.353 | |
| SuHaII | ABS | 77.548 | 70.363 | t = 3.960 | 0.000 |
| ALL | 86.828 | 79.532 | t = 3.630 | 0.000 | |
| DQ | ABS | 57.961 | 50.322 | Z = −4.604 | 0.000 |
| ALL | 61.375 | 53.769 | Z = −4.435 | 0.000 | |
| DR | ABS | 46.879 | 44.284 | t = 2.298 | 0.024 |
| ALL | 55.732 | 52.649 | t = 2.235 | 0.027 |
| ABS | ALL | |||
|---|---|---|---|---|
| F | P | F | P | |
| SuHa | 9.004 | 0.003 | 8.518 | 0.004 |
| SuHaI | 1.241 | 0.266 | 0.919 | 0.338 |
| SuHaII | 10.933 | 0.001 | 8.634 | 0.003 |
| DQ | 11.649 | 0.001 | 10.11 | 0.002 |
| DR | 3.33 | 0.068 | 2.861 | 0.091 |
| C | 3.849 | 0.050 | 3.498 | 0.062 |
| I | 0.406 | 0.524 | 0.475 | 0.491 |
| L | 0.122 | 0.727 | 0.469 | 0.494 |
| DQA1 | 10.915 | 0.001 | 9,465 | 0.002 |
| DQA2 | 4.688 | 0.031 | 4.688 | 0.031 |
| DQB1 | 6.953 | 0.009 | 6.88 | 0.009 |
| DRB3 | 3.33 | 0.068 | 2.861 | 0.091 |
| Locus | Observed Zygotes | Other Zygotes | Statistics | P | |
|---|---|---|---|---|---|
| SuHa | ABS | 31.119 | 31.153 | Z = −1.184 | 0.237 |
| ALL | 34.056 | 34.383 | t = −0.228 | 0.820 | |
| SuHaI | ABS | 22.290 | 23.649 | Z = −0.096 | 0.924 |
| ALL | 24.082 | 25.684 | Z = −0.027 | 0.978 | |
| SuHaII | ABS | 19.959 | 18.785 | Z = −1.696 | 0.090 |
| ALL | 22.136 | 21.261 | Z = −1.108 | 0.268 | |
| DQ | ABS | 15.164 | 13.717 | Z = −2.624 | 0.009 |
| ALL | 15.997 | 14.677 | Z = −2.526 | 0.012 | |
| DR | ABS | 11.857 | 11.633 | Z = −0.304 | 0.761 |
| ALL | 13.924 | 13.943 | Z = −0.023 | 0.982 |
| ABS | ALL | ||||
|---|---|---|---|---|---|
| df | F | P | F | P | |
| SuHa | 472 | 0.671 | 0.413 | 0.989 | 0.320 |
| SuHaI | 467 | 1.827 | 0.177 | 2.072 | 0.151 |
| SuHaII | 453 | 0.531 | 0.466 | 0.120 | 0.730 |
| DQ | 453 | 0.001 | 0.982 | 12.32 | 0.000 |
| DR | 344 | 0.007 | 0.935 | 0.101 | 0.751 |
| C | 283 | 0.205 | 0.651 | 0.135 | 0.714 |
| I | 256 | 5.440 | 0.020 | 5.749 | 0.017 |
| L | 252 | 0.902 | 0.343 | 1.136 | 0.288 |
| DQA1 | 399 | 7.058 | 0.008 | 6.325 | 0.012 |
| DQA2 | 146 | 16.090 | 0.000 | 16.09 | 0.000 |
| DQB1 | 242 | 0.107 | 0.743 | 0.072 | 0.788 |
| DRB3 | 344 | 0.007 | 0.935 | 0.101 | 0.751 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Wan, Q.-H.; Zhang, H.-M.; Fang, S.-G. Reproductive Strategy Inferred from Major Histocompatibility Complex-Based Inter-Individual, Sperm-Egg, and Mother-Fetus Recognitions in Giant Pandas (Ailuropoda melanoleuca). Cells 2019, 8, 257. https://doi.org/10.3390/cells8030257
Zhu Y, Wan Q-H, Zhang H-M, Fang S-G. Reproductive Strategy Inferred from Major Histocompatibility Complex-Based Inter-Individual, Sperm-Egg, and Mother-Fetus Recognitions in Giant Pandas (Ailuropoda melanoleuca). Cells. 2019; 8(3):257. https://doi.org/10.3390/cells8030257
Chicago/Turabian StyleZhu, Ying, Qiu-Hong Wan, He-Min Zhang, and Sheng-Guo Fang. 2019. "Reproductive Strategy Inferred from Major Histocompatibility Complex-Based Inter-Individual, Sperm-Egg, and Mother-Fetus Recognitions in Giant Pandas (Ailuropoda melanoleuca)" Cells 8, no. 3: 257. https://doi.org/10.3390/cells8030257
APA StyleZhu, Y., Wan, Q. -H., Zhang, H. -M., & Fang, S. -G. (2019). Reproductive Strategy Inferred from Major Histocompatibility Complex-Based Inter-Individual, Sperm-Egg, and Mother-Fetus Recognitions in Giant Pandas (Ailuropoda melanoleuca). Cells, 8(3), 257. https://doi.org/10.3390/cells8030257