The Copper(II)-Assisted Connection between NGF and BDNF by Means of Nerve Growth Factor-Mimicking Short Peptides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Simulation Details
2.1.1. Parallel Tempering Simulations
2.1.2. Docking Simulations
2.2. Peptide Synthesis
2.3. Spectroscopic Measurements
2.3.1. UV-Visible Measurements
2.3.2. Circular Dichroism (CD) Measurements
2.4. Cell Experiments
2.4.1. Materials
2.4.2. Rat Pheochromocytoma Cultures
2.4.3. Animals
2.4.4. Septal Neurons Primary Cultures
2.4.5. Proliferation Assay
2.4.6. Neurite Growth Assay
2.4.7. Protein Cellular Lysates Preparation
2.4.8. SDS-PAGE, Western Blot Analysis and Densitometry
2.4.9. Internalization of TrkA
2.4.10. Enzyme Linked Immuno Sorbent Assay (ELISA)
2.4.11. Confocal Microscopy Imaging
2.5. Data Analysis
3. Results and Discussions
3.1. Computation Studies Reveal that NGF(1-14), but not NGF(14-1) nor sNGF(1-14) Interact with TrkA-D5
3.2. The Sequence-Dependent Interaction of Dimeric Peptide d-NGF(1-15) with Domain 5 of TrkA Is Greater than that of NGF(1-14)
3.3. Cu2+ Interaction with d-NGF(1-15) Is Similar and Slightly Stronger than that with NGF(1-14), but Significantly Different Compared with that With s-NGF(1-14) and NGF(14-1)
3.4. d-NGF(1-15), NGF(1-14), and NGF Affect Proliferation and Morphology/Differentiation of PC12 Cells Conversely s-NGF(1-14) and NGF(14-1) Do Not
3.5. The d-NGF(1-15) Is a Better TrkA (Y490) Activator than NGF(1-14), while s-NGF(1-14) and NGF(14-1) Do Not Induce Any Significant Effect
3.6. d-NGF(1-15) Induces TrkA Receptor Internalization in PC12 Cells only in the Presence of CuSO4 Whereas Does Not Affect p75 Receptor Internalization
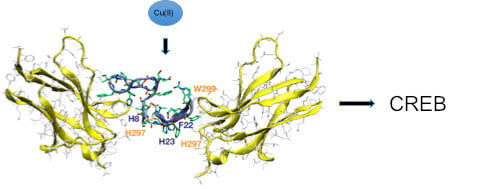
3.7. d-NGF(1–15) Signaling via TrkA Leads to Activation of cAMP Response Element-Binding Protein
3.8. Copper(II) Ion, NGF(1-14) and d-NGF(1-15) Induce BDNF Secretion
3.9. NGF(1-14) and d-NGF(1-15) Inhibits Protein Tyrosine-Phosphatase Activity
3.10. NGF(1-14) and d-NGF(1-15) Peptides Display Different Trafficking and Ionophore Activity in PC12 Cells
3.11. NGF1-14 Retains the Ability of the Full-Length Wild-Type NGF in Suppressing the Degeneration of Primary NGF-Target Neurons
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Skaper, S.D. The Neurotrophin Family of Neurotrophic Factors: An Overview. In Neurotrophic Factors; Humana Press: New York, NY, USA, 2012; pp. 1–12. [Google Scholar]
- Bothwell, M. NGF, BDNF, NT3, and NT4. In Neurotrophic Factors; Springer: Berlin/Heidelberg, Germany, 2014; pp. 3–15. [Google Scholar]
- Barbacid, M. Neurotrophic factors and their receptors. Curr. Opin. Cell Biol. 1995, 7, 148–155. [Google Scholar] [CrossRef]
- Ullrich, A.; Schlessinger, J. Signal transduction by receptors with tyrosine kinase activity. Cell 1990, 61, 203–212. [Google Scholar] [CrossRef]
- Underwood, C.K.; Coulson, E.J. The p75 neurotrophin receptor. Int. J. Biochem. Cell Biol. 2008, 40, 1664–1668. [Google Scholar] [CrossRef]
- Cohen, S.; Levi-Montalcini, R.; Hamburger, V. A Nerve Growth-Stimulating Factor Isolated from Sarcom as 37 and 180. Proc. Natl. Acad. Sci. USA 1954, 40, 1014–1018. [Google Scholar] [CrossRef] [PubMed]
- Berg, M.M.; Sternberg, D.W.; Hempstead, B.L.; Chao, M.V. The low-affinity p75 nerve growth factor (NGF) receptor mediates NGF-induced tyrosine phosphorylation. Proc. Natl. Acad. Sci. USA 1991, 88, 7106–7110. [Google Scholar] [CrossRef]
- Chao, M.V. Neurotrophins and their receptors: A convergence point for many signaling pathways. Nat. Rev. Neurosci. 2003, 4, 299–309. [Google Scholar] [CrossRef]
- Schneider, R.; Schweiger, M. A novel modular mosaic of cell adhesion motifs in the extracellular domains of the neurogenic trk and trkB tyrosine kinase receptors. Oncogene 1991, 6, 1807–1811. [Google Scholar] [PubMed]
- Blum, R.; Konnerth, A. Neurotrophin-Mediated Rapid Signaling in the Central Nervous System: Mechanisms and Functions. Physiology 2005, 20, 70–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaccaro, M.C.; Ivanisevic, L.; Perez, P.; Meakin, S.O.; Saragovi, H.U. p75 Co-receptors Regulate Ligand-dependent and Ligand-independent Trk Receptor Activation, in Part by Altering Trk Docking Subdomains. J. Biol. Chem. 2001, 276, 31023–31029. [Google Scholar] [CrossRef]
- Skaper, S.D. The biology of neurotrophins, signaling pathways, and functional peptide mimetics of neurotrophins and their receptors. CNS Neurol. Disord. Drug Targets 2008, 7, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Peleshok, J.; Saragovi, H.U. Functional mimetics of neurotrophins and their receptors. Biochem. Soc. Trans. 2006, 34, 612–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saragovi, H.U.; Gehring, K. Development of pharmacological agents for targeting neurotrophins and their receptors. Trends Pharmacol. Sci. 2000, 21, 93–98. [Google Scholar] [CrossRef]
- Longo, F.M.; Massa, S.M. Small-molecule modulation of neurotrophin receptors: A strategy for the treatment of neurological disease. Nat. Rev. Drug Discov. 2013, 12, 507–525. [Google Scholar] [CrossRef] [PubMed]
- Kazim, S.F.; Iqbal, K. Neurotrophic factor small-molecule mimetics mediated neuroregeneration and synaptic repair: Emerging therapeutic modality for Alzheimer’s disease. Mol. Neurodegener. 2016, 11. [Google Scholar] [CrossRef]
- Josephy-Hernandez, S.; Jmaeff, S.; Pirvulescu, I.; Aboulkassim, T.; Saragovi, H.U. Neurotrophin receptor agonists and antagonists as therapeutic agents: An evolving paradigm. Neurobiol. Dis. 2017, 97, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Coulson, E.J.; Simmons, D.A.; Knowles, J.K.; Belichenko, N.P.; Banerjee, G.; Finkle, C.; Massa, S.M.; Longo, F.M. A Small Molecule p75NTR Ligand, LM11A-31, Reverses Cholinergic Neurite Dystrophy in Alzheimer’s Disease Mouse Models with Mid- to Late-Stage Disease Progression. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Habtemariam, S. The brain-derived neurotrophic factor in neuronal plasticity and neuroregeneration: New pharmacological concepts for old and new drugs. Neural Regen. Res. 2018, 13. [Google Scholar] [CrossRef] [PubMed]
- Travaglia, A.; Pietropaolo, A.; Di Martino, R.; Nicoletti, V.G.; La Mendola, D.; Calissano, P.; Rizzarelli, E. A Small Linear Peptide Encompassing the NGF N-Terminus Partly Mimics the Biological Activities of the Entire Neurotrophin in PC12 Cells. ACS Chem. Neurosci. 2015, 6, 1379–1392. [Google Scholar] [CrossRef]
- Pandini, G.; Satriano, C.; Pietropaolo, A.; Gianì, F.; Travaglia, A.; La Mendola, D.; Nicoletti, V.G.; Rizzarelli, E. The Inorganic Side of NGF: Copper(II) and Zinc(II) Affect the NGF Mimicking Signaling of the N-Terminus Peptides Encompassing the Recognition Domain of TrkA Receptor. Front. Neurosci. 2016, 10. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S.; Tavazoie, S.F.; Maloratsky, A.; Jacobs, K.M.; Harris, K.M.; Greenberg, M.E. CREB: A major mediator of neuronal neurotrophin responses. Neuron 1997, 19, 1031–1047. [Google Scholar] [CrossRef]
- Travaglia, A.; Pietropaolo, A.; La Mendola, D.; Nicoletti, V.G.; Rizzarelli, E. The inorganic perspectives of neurotrophins and Alzheimer’s disease. J. Inorg. Biochem. 2012, 111, 130–137. [Google Scholar] [CrossRef]
- Kheirvari, S.; Uezu, K.; Yamamoto, S.; Nakaya, Y. High-dose dietary supplementation of vitamin A induces brain-derived neurotrophic factor and nerve growth factor production in mice with simultaneous deficiency of vitamin A and zinc. Nutr. Neurosci. 2008, 11, 228–234. [Google Scholar] [CrossRef]
- Allington, C.; Shamovsky, I.L.; Ross, G.M.; Riopelle, R.J. Zinc inhibits p75NTR-mediated apoptosis in chick neural retina. Cell Death Differ. 2001, 8, 451–456. [Google Scholar] [CrossRef] [Green Version]
- Birkaya, B.; Aletta, J.M. NGF promotes copper accumulation required for optimum neurite outgrowth and protein methylation. J. Neurobiol. 2005, 63, 49–61. [Google Scholar] [CrossRef] [Green Version]
- Bica, L.; Liddell, J.R.; Donnelly, P.S.; Duncan, C.; Caragounis, A.; Volitakis, I.; Paterson, B.M.; Cappai, R.; Grubman, A.; Camakaris, J.; et al. Neuroprotective copper bis(thiosemicarbazonato) complexes promote neurite elongation. PLoS ONE 2014, 9, e90070. [Google Scholar] [CrossRef]
- Ross, G.M.; Shamovsky, I.L.; Lawrance, G.; Solc, M.; Dostaler, S.M.; Jimmo, S.L.; Weaver, D.F.; Riopelle, R.J. Zinc alters conformation and inhibits biological activities of nerve growth factor and related neurotrophins. Nat. Med. 1997, 3, 872–878. [Google Scholar] [CrossRef]
- Maitra, R.; Shamovsky, I.L.; Wang, W.; Solc, M.; Lawrance, G.; Dostaler, S.M.; Ross, G.M.; Riopelle, R.J. Differential effects of transition metal cations on the conformation and biological activities of nerve growth factor. Neurotox. Res. 2000, 2, 321–341. [Google Scholar] [CrossRef]
- Wang, J.K. Cu2+ induces Ca2+-dependent neurotransmitter release from brain catecholaminergic nerve terminals. Eur. J. Pharmacol. 1999, 373, 163–169. [Google Scholar] [CrossRef]
- Maliartchouk, S.; Debeir, T.; Beglova, N.; Cuello, A.C.; Gehring, K.; Saragovi, H.U. Genuine monovalent ligands of TrkA nerve growth factor receptors reveal a novel pharmacological mechanism of action. J. Biol. Chem. 2000, 275, 9946–9956. [Google Scholar] [CrossRef]
- Colangelo, A.M.; Bianco, M.R.; Vitagliano, L.; Cavaliere, C.; Cirillo, G.; De Gioia, L.; Diana, D.; Colombo, D.; Redaelli, C.; Zaccaro, L.; et al. A new nerve growth factor-mimetic peptide active on neuropathic pain in rats. J. Neurosci. 2008, 28, 2698–2709. [Google Scholar] [CrossRef]
- Marte, A.; Messa, M.; Benfenati, F.; Onofri, F. Synapsins Are Downstream Players of the BDNF-Mediated Axonal Growth. Mol. Neurobiol. 2016, 54, 484–494. [Google Scholar] [CrossRef]
- Peng, S.; Li, W.; Lv, L.; Zhang, Z.; Zhan, X. BDNF as a biomarker in diagnosis and evaluation of treatment for schizophrenia and depression. Discov. Med. 2018, 26, 127–136. [Google Scholar]
- Yang, Y.; Liu, Y.; Wang, G.; Hei, G.; Wang, X.; Li, R.; Li, L.; Wu, R.; Zhao, J. Brain-derived neurotrophic factor is associated with cognitive impairments in first-episode and chronic schizophrenia. Psychiatry Res. 2019, 273, 528–536. [Google Scholar] [CrossRef]
- Notaras, M.; Hill, R.; Gogos, J.A.; van den Buuse, M. BDNF Val66Met genotype determines hippocampus-dependent behavior via sensitivity to glucocorticoid signaling. Mol. Psychiatry 2015, 21, 730–732. [Google Scholar] [CrossRef]
- Beck, M.; Flachenecker, P.; Magnus, T.; Giess, R.; Reiners, K.; Toyka, K.V.; Naumann, M. Autonomic dysfunction in ALS: A preliminary study on the effects of intrathecal BDNF. Amyotroph. Lateral Scler. 2009, 6, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct. Funct. Bioinform. 2006, 65, 712–725. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Rosta, E.; Buchete, N.V.; Hummer, G. Thermostat artifacts in replica exchange molecular dynamics simulations. J. Chem. Theory Comput. 2009, 5, 1393–1399. [Google Scholar] [CrossRef] [Green Version]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide Folding: When Simulation Meets Experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Wiesmann, C.; Ultsch, M.H.; Bass, S.H.; de Vos, A.M. Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature 1999, 401, 184–188. [Google Scholar] [CrossRef]
- De Vries, S.J.; van Dijk, M.; Bonvin, A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010, 5, 883–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Travaglia, A.; Arena, G.; Fattorusso, R.; Isernia, C.; La Mendola, D.; Malgieri, G.; Nicoletti, V.G.; Rizzarelli, E. The Inorganic Perspective of Nerve Growth Factor: Interactions of Cu2+ and Zn2+ with the N-Terminus Fragment of Nerve Growth Factor Encompassing the Recognition Domain of the TrkA Receptor. Chem. - A Eur. J. 2011, 17, 3726–3738. [Google Scholar] [CrossRef]
- Forte, G.; Travaglia, A.; Magri, A.; Satriano, C.; La Mendola, D. Adsorption of NGF and BDNF derived peptides on gold surfaces. Phys. Chem. Chem. Phys. 2014, 16, 1536–1544. [Google Scholar] [CrossRef]
- La Mendola, D.; Farkas, D.; Bellia, F.; Magri, A.; Travaglia, A.; Hansson, O.; Rizzarelli, E. Probing the copper(II) binding features of angiogenin. Similarities and differences between a N-terminus peptide fragment and the recombinant human protein. Inorg. Chem. 2012, 51, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Latina, V.; Caioli, S.; Zona, C.; Ciotti, M.T.; Amadoro, G.; Calissano, P. Impaired NGF/TrkA Signaling Causes Early AD-Linked Presynaptic Dysfunction in Cholinergic Primary Neurons. Front. Cell. Neurosci. 2017, 11. [Google Scholar] [CrossRef] [PubMed]
- Troncone, G.; Walter, R.F.H.; Werner, R.; Vollbrecht, C.; Hager, T.; Flom, E.; Christoph, D.C.; Schmeller, J.; Schmid, K.W.; Wohlschlaeger, J.; et al. ACTB, CDKN1B, GAPDH, GRB2, RHOA and SDCBP Were Identified as Reference Genes in Neuroendocrine Lung Cancer via the nCounter Technology. Plos ONE 2016, 11. [Google Scholar] [CrossRef]
- Miller, E.W.; Zeng, L.; Domaille, D.W.; Chang, C.J. Preparation and use of Coppersensor-1, a synthetic fluorophore for live-cell copper imaging. Nat. Protoc. 2006, 1, 824–827. [Google Scholar] [CrossRef]
- Settanni, G.; Cattaneo, A.; Carloni, P. Molecular Dynamics Simulations of the NGF-TrkA Domain 5 Complex and Comparison with Biological Data. Biophys. J. 2003, 84, 2282–2292. [Google Scholar] [CrossRef] [Green Version]
- Wehrman, T.; He, X.; Raab, B.; Dukipatti, A.; Blau, H.; Garcia, K.C. Structural and Mechanistic Insights into Nerve Growth Factor Interactions with the TrkA and p75 Receptors. Neuron 2007, 53, 25–38. [Google Scholar] [CrossRef] [Green Version]
- Lessmann, V.; Gottmann, K.; Malcangio, M. Neurotrophin secretion: Current facts and future prospects. Prog. Neurobiol. 2003, 69, 341–374. [Google Scholar] [CrossRef]
- Scarpi, D.; Cirelli, D.; Matrone, C.; Castronovo, G.; Rosini, P.; Occhiato, E.G.; Romano, F.; Bartali, L.; Clemente, A.M.; Bottegoni, G.; et al. Low molecular weight, non-peptidic agonists of TrkA receptor with NGF-mimetic activity. Cell Death Dis. 2012, 3, e339. [Google Scholar] [CrossRef]
- Satriano, C.; Forte, G.; Magri, A.; Di Pietro, P.; Travaglia, A.; Pandini, G.; Giani, F.; La Mendola, D. Neurotrophin-mimicking peptides at the biointerface with gold respond to copper ion stimuli. Phys. Chem. Chem. Phys. 2016, 18, 30595–30604. [Google Scholar] [CrossRef]
- Sóvágó, I.; Ősz, K. Metal ion selectivity of oligopeptides. Dalton Trans. 2006, 3841–3854. [Google Scholar] [CrossRef]
- La Mendola, D.; Arnesano, F.; Hansson, O.; Giacomelli, C.; Calo, V.; Mangini, V.; Magri, A.; Bellia, F.; Trincavelli, M.L.; Martini, C.; et al. Copper binding to naturally occurring, lactam form of angiogenin differs from that to recombinant protein, affecting their activity. Metallomics 2016, 8, 118–124. [Google Scholar] [CrossRef]
- La Mendola, D.; Magri, A.; Campagna, T.; Campitiello, M.A.; Raiola, L.; Isernia, C.; Hansson, O.; Bonomo, R.P.; Rizzarelli, E. A doppel alpha-helix peptide fragment mimics the copper(II) interactions with the whole protein. Chemistry 2010, 16, 6212–6223. [Google Scholar] [CrossRef]
- La Mendola, D.; Magri, A.; Santoro, A.M.; Nicoletti, V.G.; Rizzarelli, E. Copper(II) interaction with peptide fragments of histidine-proline-rich glycoprotein: Speciation, stability and binding details. J. Inorg Biochem 2012, 111, 59–69. [Google Scholar] [CrossRef]
- Bellia, F.; La Mendola, D.; Maccarrone, G.; Mineo, P.; Vitalini, D.; Scamporrino, E.; Sortino, S.; Vecchio, G.; Rizzarelli, E. Copper(II) complexes with β-cyclodextrin–homocarnosine conjugates and their antioxidant activity. Inorg. Chim. Acta 2007, 360, 945–954. [Google Scholar] [CrossRef]
- Ross, G.M.; Shamovsky, I.L.; Woo, S.B.; Post, J.I.; Vrkljan, P.N.; Lawrance, G.; Solc, M.; Dostaler, S.M.; Neet, K.E.; Riopelle, R.J. The binding of zinc and copper ions to nerve growth factor is differentially affected by pH: Implications for cerebral acidosis. J. Neurochem. 2001, 78, 515–523. [Google Scholar] [CrossRef]
- White, A.R.; Barnham, K.J.; Huang, X.; Voltakis, I.; Beyreuther, K.; Masters, C.L.; Cherny, R.A.; Bush, A.I.; Cappai, R. Iron inhibits neurotoxicity induced by trace copper and biological reductants. J. Biol. Inorg. Chem. 2004, 9, 269–280. [Google Scholar] [CrossRef]
- Naletova, I.; Satriano, C.; Curci, A.; Margiotta, N.; Natile, G.; Arena, G.; La Mendola, D.; Nicoletti, V.G.; Rizzarelli, E. Cytotoxic phenanthroline derivatives alter metallostasis and redox homeostasis in neuroblastoma cells. Oncotarget 2018, 9, 36289–36316. [Google Scholar] [CrossRef]
- Reichardt, L.F. Neurotrophin-regulated signaling pathways. Philos Trans. R Soc. Lond. B Biol. Sci. 2006, 361, 1545–1564. [Google Scholar] [CrossRef]
- Van der Heide, L.P.; Ramakers, G.M.; Smidt, M.P. Insulin signaling in the central nervous system: Learning to survive. Prog. Neurobiol. 2006, 79, 205–221. [Google Scholar] [CrossRef]
- Sweatt, J.D. Mitogen-activated protein kinases in synaptic plasticity and memory. Curr. Opin. Neurobiol. 2004, 14, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Impey, S.; McCorkle, S.R.; Cha-Molstad, H.; Dwyer, J.M.; Yochum, G.S.; Boss, J.M.; McWeeney, S.; Dunn, J.J.; Mandel, G.; Goodman, R.H. Defining the CREB regulon: A genome-wide analysis of transcription factor regulatory regions. Cell 2004, 119, 1041–1054. [Google Scholar] [CrossRef]
- Barco, A.; Bailey, C.H.; Kandel, E.R. Common molecular mechanisms in explicit and implicit memory. J. Neurochem. 2006, 97, 1520–1533. [Google Scholar] [CrossRef] [Green Version]
- Abel, T.; Nguyen, P.V. Regulation of hippocampus-dependent memory by cyclic AMP-dependent protein kinase. Prog. Brain Res. 2008, 169, 97–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowianski, P.; Lietzau, G.; Czuba, E.; Waskow, M.; Steliga, A.; Morys, J. BDNF: A Key Factor with Multipotent Impact on Brain Signaling and Synaptic Plasticity. Cell Mol. Neurobiol. 2018, 38, 579–593. [Google Scholar] [CrossRef]
- Motamedi, S.; Karimi, I.; Jafari, F. The interrelationship of metabolic syndrome and neurodegenerative diseases with focus on brain-derived neurotrophic factor (BDNF): Kill two birds with one stone. Metab. Brain Dis. 2017, 32, 651–665. [Google Scholar] [CrossRef] [PubMed]
- Tonks, N.K. Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013, 280, 346–378. [Google Scholar] [CrossRef]
- Singh, K.B.; Maret, W. The interactions of metal cations and oxyanions with protein tyrosine phosphatase 1B. Biometals 2017, 30, 517–527. [Google Scholar] [CrossRef] [Green Version]
- Trusso Sfrazzetto, G.; Satriano, C.; Tomaselli, G.A.; Rizzarelli, E. Synthetic fluorescent probes to map metallostasis and intracellular fate of zinc and copper. Coord. Chem. Rev. 2016, 311, 125–167. [Google Scholar] [CrossRef]
- Cheng, Q.; Song, S.H.; Augustine, G.J. Calcium-Dependent and Synapsin-Dependent Pathways for the Presynaptic Actions of BDNF. Front. Cell Neurosci. 2017, 11, 75. [Google Scholar] [CrossRef]
- Crouch, P.J.; Hung, L.W.; Adlard, P.A.; Cortes, M.; Lal, V.; Filiz, G.; Perez, K.A.; Nurjono, M.; Caragounis, A.; Du, T.; et al. Increasing Cu bioavailability inhibits Abeta oligomers and tau phosphorylation. Proc. Natl. Acad. Sci. USA 2009, 106, 381–386. [Google Scholar] [CrossRef]
- White, A.R.; Du, T.; Laughton, K.M.; Volitakis, I.; Sharples, R.A.; Xilinas, M.E.; Hoke, D.E.; Holsinger, R.M.; Evin, G.; Cherny, R.A.; et al. Degradation of the Alzheimer disease amyloid beta-peptide by metal-dependent up-regulation of metalloprotease activity. J. Biol. Chem. 2006, 281, 17670–17680. [Google Scholar] [CrossRef]
- Wu, W.; Samet, J.M.; Silbajoris, R.; Dailey, L.A.; Sheppard, D.; Bromberg, P.A.; Graves, L.M. Heparin-binding epidermal growth factor cleavage mediates zinc-induced epidermal growth factor receptor phosphorylation. Am. J. Respir. Cell Mol. Biol. 2004, 30, 540–547. [Google Scholar] [CrossRef]
- De Souza, A.P.; Gerlach, R.F.; Line, S.R. Inhibition of human gingival gelatinases (MMP-2 and MMP-9) by metal salts. Dent. Mater. 2000, 16, 103–108. [Google Scholar] [CrossRef]
- Hwang, J.J.; Park, M.H.; Koh, J.Y. Copper activates TrkB in cortical neurons in a metalloproteinase-dependent manner. J. Neurosci. Res. 2007, 85, 2160–2166. [Google Scholar] [CrossRef]
- Minami, A.; Takeda, A.; Yamaide, R.; Oku, N. Relationship between zinc and neurotransmitters released into the amygdalar extracellular space. Brain Res. 2002, 936, 91–94. [Google Scholar] [CrossRef]
- Hopt, A.; Korte, S.; Fink, H.; Panne, U.; Niessner, R.; Jahn, R.; Kretzschmar, H.; Herms, J. Methods for studying synaptosomal copper release. J. Neurosci. Methods 2003, 128, 159–172. [Google Scholar] [CrossRef]
- Kubiatowski, T.; Jang, T.; Lachyankar, M.B.; Salmonsen, R.; Nabi, R.R.; Quesenberry, P.J.; Litofsky, N.S.; Ross, A.H.; Recht, L.D. Association of increased phosphatidylinositol 3-kinase signaling with increased invasiveness and gelatinase activity in malignant gliomas. J. Neurosurg. 2001, 95, 480–488. [Google Scholar] [CrossRef]
- Matsumoto, K.; Minamitani, T.; Orba, Y.; Sato, M.; Sawa, H.; Ariga, H. Induction of matrix metalloproteinase-2 by tenascin-X deficiency is mediated through the c-Jun N-terminal kinase and protein tyrosine kinase phosphorylation pathway. Exp. Cell Res. 2004, 297, 404–414. [Google Scholar] [CrossRef]
- Iulita, M.F.; Bistue Millon, M.B.; Pentz, R.; Aguilar, L.F.; Do Carmo, S.; Allard, S.; Michalski, B.; Wilson, E.N.; Ducatenzeiler, A.; Bruno, M.A.; et al. Differential deregulation of NGF and BDNF neurotrophins in a transgenic rat model of Alzheimer’s disease. Neurobiol. Dis. 2017, 108, 307–323. [Google Scholar] [CrossRef]
- Burns, A.; Iliffe, S. Alzheimer’s disease. BMJ 2009, 338, b158. [Google Scholar] [CrossRef] [PubMed]
- Francis, B.M.; Kim, J.; Barakat, M.E.; Fraenkl, S.; Yucel, Y.H.; Peng, S.; Michalski, B.; Fahnestock, M.; McLaurin, J.; Mount, H.T. Object recognition memory and BDNF expression are reduced in young TgCRND8 mice. Neurobiol. Aging 2012, 33, 555–563. [Google Scholar] [CrossRef]
- Hock, C.; Heese, K.; Hulette, C.; Rosenberg, C.; Otten, U. Region-specific neurotrophin imbalances in Alzheimer disease: Decreased levels of brain-derived neurotrophic factor and increased levels of nerve growth factor in hippocampus and cortical areas. Arch. Neurol. 2000, 57, 846–851. [Google Scholar] [CrossRef]
- Connor, B.; Young, D.; Yan, Q.; Faull, R.L.; Synek, B.; Dragunow, M. Brain-derived neurotrophic factor is reduced in Alzheimer’s disease. Mol. Brain Res. 1997, 49, 71–81. [Google Scholar] [CrossRef]
- Choi, S.H.; Bylykbashi, E.; Chatila, Z.K.; Lee, S.W.; Pulli, B.; Clemenson, G.D.; Kim, E.; Rompala, A.; Oram, M.K.; Asselin, C.; et al. Combined adult neurogenesis and BDNF mimic exercise effects on cognition in an Alzheimer’s mouse model. Science 2018, 361. [Google Scholar] [CrossRef] [PubMed]












 ,
,  ) or copper-supplemented medium (
) or copper-supplemented medium (  ,
,  ). In (D,E): quantitative analyses of total intracellular copper (D) or nuclear-confined copper (E) after PC12 cells incubation for 10, 15, 20 or 30 min with 10 μM NGF(1-14) (cytoplasm:
). In (D,E): quantitative analyses of total intracellular copper (D) or nuclear-confined copper (E) after PC12 cells incubation for 10, 15, 20 or 30 min with 10 μM NGF(1-14) (cytoplasm:  ; nuclei:
; nuclei:  ) or 10 μM d-NGF(1-14) (cytoplasm:
) or 10 μM d-NGF(1-14) (cytoplasm:  ; nuclei:
; nuclei:  ). ** p < 0.01, *** p < 0.001, versus control.; ** p < 0.01, *** p < 0.001, **** p < 0.0001 versus the preceding incubation time; § p < 0.05, §§§ p < 0.001 versus [NGF(1-14)].
). ** p < 0.01, *** p < 0.001, versus control.; ** p < 0.01, *** p < 0.001, **** p < 0.0001 versus the preceding incubation time; § p < 0.05, §§§ p < 0.001 versus [NGF(1-14)].
 ,
,  ) or copper-supplemented medium (
) or copper-supplemented medium (  ,
,  ). In (D,E): quantitative analyses of total intracellular copper (D) or nuclear-confined copper (E) after PC12 cells incubation for 10, 15, 20 or 30 min with 10 μM NGF(1-14) (cytoplasm:
). In (D,E): quantitative analyses of total intracellular copper (D) or nuclear-confined copper (E) after PC12 cells incubation for 10, 15, 20 or 30 min with 10 μM NGF(1-14) (cytoplasm:  ; nuclei:
; nuclei:  ) or 10 μM d-NGF(1-14) (cytoplasm:
) or 10 μM d-NGF(1-14) (cytoplasm:  ; nuclei:
; nuclei:  ). ** p < 0.01, *** p < 0.001, versus control.; ** p < 0.01, *** p < 0.001, **** p < 0.0001 versus the preceding incubation time; § p < 0.05, §§§ p < 0.001 versus [NGF(1-14)].
). ** p < 0.01, *** p < 0.001, versus control.; ** p < 0.01, *** p < 0.001, **** p < 0.0001 versus the preceding incubation time; § p < 0.05, §§§ p < 0.001 versus [NGF(1-14)].
| Peptide | λ/nm (ε/M−1 cm−1) | λ/nm (Δε/M−1 cm−1) |
|---|---|---|
| NGF(1-14) a | 605 (104) | 284 (−1.40), 328 (0.55), 340 (0.22), 632 (−0.58) |
| d-NGF(1-15) | 590 (130) | 288 (−3.18), 322 (1.15), 500 (0.11), 587 (−0.58) |
| s-NGF(1-14) | 618 (85) | 288 (−1.150), 324 (0.70), 599 (−0.57) |
| NGF(14-1) | 615 (120) | 282 (−0.42), 316 (−0.32), 354 (0.07); 515 (0.31), 626 (−0,62) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naletova, I.; Satriano, C.; Pietropaolo, A.; Gianì, F.; Pandini, G.; Triaca, V.; Amadoro, G.; Latina, V.; Calissano, P.; Travaglia, A.; et al. The Copper(II)-Assisted Connection between NGF and BDNF by Means of Nerve Growth Factor-Mimicking Short Peptides. Cells 2019, 8, 301. https://doi.org/10.3390/cells8040301
Naletova I, Satriano C, Pietropaolo A, Gianì F, Pandini G, Triaca V, Amadoro G, Latina V, Calissano P, Travaglia A, et al. The Copper(II)-Assisted Connection between NGF and BDNF by Means of Nerve Growth Factor-Mimicking Short Peptides. Cells. 2019; 8(4):301. https://doi.org/10.3390/cells8040301
Chicago/Turabian StyleNaletova, Irina, Cristina Satriano, Adriana Pietropaolo, Fiorenza Gianì, Giuseppe Pandini, Viviana Triaca, Giuseppina Amadoro, Valentina Latina, Pietro Calissano, Alessio Travaglia, and et al. 2019. "The Copper(II)-Assisted Connection between NGF and BDNF by Means of Nerve Growth Factor-Mimicking Short Peptides" Cells 8, no. 4: 301. https://doi.org/10.3390/cells8040301
APA StyleNaletova, I., Satriano, C., Pietropaolo, A., Gianì, F., Pandini, G., Triaca, V., Amadoro, G., Latina, V., Calissano, P., Travaglia, A., Nicoletti, V. G., La Mendola, D., & Rizzarelli, E. (2019). The Copper(II)-Assisted Connection between NGF and BDNF by Means of Nerve Growth Factor-Mimicking Short Peptides. Cells, 8(4), 301. https://doi.org/10.3390/cells8040301