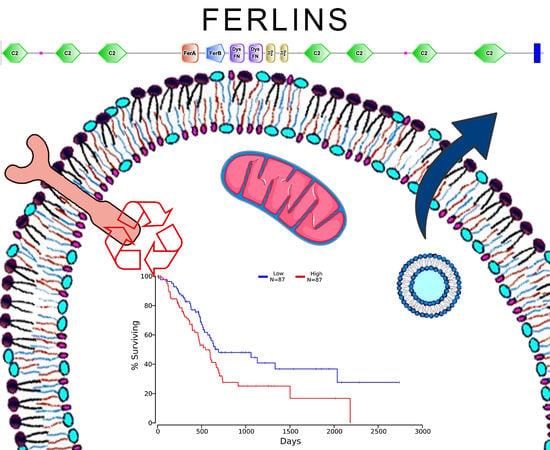
Ferlin Overview: From Membrane to Cancer Biology
Abstract
:1. Introduction
2. Genomic Organization of Ferlin Gene Family
3. Ferlin’s Structure and Localization
4. Ferlin’s Interactions with Phospholipids
5. Ferlin’s Main Functions in Non-Neoplastic Cells and Tissues
5.1. In Mammal Muscle Cells
5.2. In mammal Inner Ear Cells
5.3. In Mammal Endothelial Cells
5.4. Other Mammal’s Cells
6. Ferlins in Cancer, Potential Targets to Kill Cancer
6.1. Breast Cancer and Melanoma
6.2. Pancreas and Colon Cancers
6.3. Lung Cancer
6.4. Liver Cancer
6.5. Head and Neck Cancer
6.6. Gastric Cancer
6.7. Gynecological Cancers
7. Conclusions
8. Statistical Methods
Funding
Acknowledgments
Conflicts of Interest
References
- Bernardes, N.; Fialho, A.M. Perturbing the Dynamics and Organization of Cell Membrane Components: A New Paradigm for Cancer-Targeted Therapies. Int. J. Mol. Sci. 2018, 19, 3871. [Google Scholar] [CrossRef] [PubMed]
- Achanzar, W.E.; Ward, S. A nematode gene required for sperm vesicle fusion. J. Cell Sci. 1997, 110, 1073–1081. [Google Scholar] [PubMed]
- Bashir, R.; Britton, S.; Strachan, T.; Keers, S.; Vafiadaki, E.; Lako, M.; Richard, I.; Marchand, S.; Bourg, N.; Argov, Z.; et al. A gene related to caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat. Genet. 1998, 20, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Yasunaga, S.; Grati, M.; Cohen-Salmon, M.; El-Amraoui, A.; Mustapha, M.; Salem, N.; El-Zir, E.; Loiselet, J.; Petit, C. A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nat. Genet. 1999, 21, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.B.; Delmonte, A.J.; Ly, C.T.; McNally, E.M. Myoferlin, a candidate gene and potential modifier of muscular dystrophy. Hum. Mol. Genet. 2000, 9, 217–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Britton, S.; Freeman, T.; Vafiadaki, E.; Keers, S.; Harrison, R.; Bushby, K.; Bashir, R. The third human FER-1-like protein is highly similar to dysferlin. Genomics 2000, 68, 313–321. [Google Scholar] [CrossRef]
- Liu, J.; Aoki, M.; Illa, I.; Wu, C.; Fardeau, M.; Angelini, C.; Serrano, C.; Urtizberea, J.A.; Hentati, F.; Hamida, M.B.; et al. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 1998, 20, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.Y.; Ahmed, Z.M.; Riazuddin, S.; Bhinder, M.A.; Shahzad, M.; Husnain, T.; Griffith, A.J.; Friedman, T.B. Identities and frequencies of mutations of the otoferlin gene (OTOF) causing DFNB9 deafness in Pakistan. Clin. Genet. 2009, 75, 237–243. [Google Scholar] [CrossRef]
- Tekin, M.; Akcayoz, D.; Incesulu, A. A novel missense mutation in a C2 domain of OTOF results in autosomal recessive auditory neuropathy. Am. J. Med. Genet. A 2005, 138, 6–10. [Google Scholar] [CrossRef]
- Doherty, K.R.; Cave, A.; Davis, D.B.; Delmonte, A.J.; Posey, A.; Earley, J.U.; Hadhazy, M.; McNally, E.M. Normal myoblast fusion requires myoferlin. Development 2005, 132, 5565–5575. [Google Scholar] [CrossRef] [Green Version]
- Kiselev, A.; Vaz, R.; Knyazeva, A.; Sergushichev, A.; Dmitrieva, R.; Khudiakov, A.; Jorholt, J.; Smolina, N.; Sukhareva, K.; Fomicheva, Y.; et al. Truncating variant in myof gene is associated with limb-girdle type muscular dystrophy and cardiomyopathy. Front. Genet. 2019, 10, 608. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.; Liu, J.; Richard, I.; Bashir, R.; Britton, S.; Keers, S.M.; Oeltjen, J.; Brown, H.E.; Marchand, S.; Bourg, N.; et al. Genomic organization of the dysferlin gene and novel mutations in Miyoshi myopathy. Neurology 2001, 57, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Demonbreun, A.R.; Lapidos, K.A.; Heretis, K.; Levin, S.; Dale, R.; Pytel, P.; Svensson, E.C.; McNally, E.M. Myoferlin regulation by NFAT in muscle injury, regeneration and repair. J. Cell Sci. 2010, 123, 2413–2422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Corbalan-Garcia, S.; Gómez-Fernández, J.C. Signaling through C2 domains: More than one lipid target. Biochim. Biophys. Acta 2014, 1838, 1536–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Aravind, L. Identification of novel families and classification of the C2 domain superfamily elucidate the origin and evolution of membrane targeting activities in eukaryotes. Gene 2010, 469, 18–30. [Google Scholar] [CrossRef] [Green Version]
- Nalefski, E.A.; Falke, J.J. The C2 domain calcium-binding motif: Structural and functional diversity. Protein Sci. 1996, 5, 2375–2390. [Google Scholar] [CrossRef] [Green Version]
- Shin, O.-H.; Han, W.; Wang, Y.; Südhof, T.C. Evolutionarily conserved multiple C2 domain proteins with two transmembrane regions (MCTPs) and unusual Ca2+ binding properties. J. Biol. Chem. 2005, 280, 1641–1651. [Google Scholar] [CrossRef]
- Min, S.-W.; Chang, W.-P.; Südhof, T.C. E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proc. Natl. Acad. Sci. USA 2007, 104, 3823–3828. [Google Scholar] [CrossRef]
- Rizo, J.; Sudhof, T.C. C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 1998, 273, 15879–15882. [Google Scholar] [CrossRef]
- Von Poser, C.; Ichtchenko, K.; Shao, X.; Rizo, J.; Sudhof, T.C. The evolutionary pressure to inactivate. A subclass of synaptotagmins with an amino acid substitution that abolishes Ca2+ binding. J. Biol. Chem. 1997, 272, 14314–14319. [Google Scholar] [CrossRef] [PubMed]
- Lek, A.; Lek, M.; North, K.N.; Cooper, S.T. Phylogenetic analysis of ferlin genes reveals ancient eukaryotic origins. BMC Evol. Biol. 2010, 10, 231. [Google Scholar] [CrossRef] [PubMed]
- Washington, N.L.; Ward, S. FER-1 regulates Ca2+-mediated membrane fusion during C. elegans spermatogenesis. J. Cell Sci. 2006, 119, 2552–2562. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Rachubinski, D.A.; Joshi, S.; Rachubinski, R.A.; Subramani, S. Dysferlin domain-containing proteins, Pex30p and Pex31p, localized to two compartments, control the number and size of oleate-induced peroxisomes in Pichia pastoris. Mol. Biol. Cell. 2008, 19, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Harris, R.; Geddes, S.M.; Strehle, E.-M.; Watson, J.D.; Bashir, R.; Bushby, K.; Driscoll, P.C.; Keep, N.H. Solution Structure of the Inner DysF Domain of Myoferlin and Implications for Limb Girdle Muscular Dystrophy Type 2B. J. Mol. Biol. 2008, 379, 981–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuson, K.; Rice, A.; Mahling, R.; Snow, A.; Nayak, K.; Shanbhogue, P.; Meyer, A.G.; Redpath, G.M.I.; Hinderliter, A.; Cooper, S.T.; et al. Alternate splicing of dysferlin C2A confers Ca2+-dependent and Ca2+-independent binding for membrane repair. Structure 2014, 22, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A.; Van Deutekom, J.C.T.; Fokkema, I.F.; Van Ommen, G.-J.B.; Den Dunnen, J.T. Entries in the Leiden Duchenne muscular dystrophy mutation database: An overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 2006, 34, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Bernatchez, P.N.; Acevedo, L.; Fernandez-Hernando, C.; Murata, T.; Chalouni, C.; Kim, J.; Erdjument-Bromage, H.; Shah, V.; Gratton, J.-P.; McNally, E.M.; et al. Myoferlin regulates vascular endothelial growth factor receptor-2 stability and function. J. Biol. Chem. 2007, 282, 30745–30753. [Google Scholar] [CrossRef]
- Miyatake, Y.; Yamano, T.; Hanayama, R. Myoferlin-Mediated Lysosomal Exocytosis Regulates Cytotoxicity by Phagocytes. J. Immunol. 2018, 201, 3051–3057. [Google Scholar] [CrossRef]
- Redpath, G.M.I.; Sophocleous, R.A.; Turnbull, L.; Whitchurch, C.B.; Cooper, S.T. Ferlins Show Tissue-Specific Expression and Segregate as Plasma Membrane/Late Endosomal or Trans-Golgi/Recycling Ferlins. Traffic 2016, 17, 245–266. [Google Scholar] [CrossRef] [Green Version]
- Davis, D.B.; Doherty, K.R.; Delmonte, A.J.; McNally, E.M. Calcium-sensitive phospholipid binding properties of normal and mutant ferlin C2 domains. J. Biol. Chem. 2002, 277, 22883–22888. [Google Scholar] [CrossRef] [PubMed]
- Bootman, M.D.; Rietdorf, K.; Hardy, H.; Dautova, Y.; Corps, E.; Pierro, C.; Stapleton, E.; Kang, E.; Proudfoot, D. Calcium Signalling and Regulation of Cell Function. In eLS; John Wiley & Sons, Ltd.: Chichester, UK, 2012; pp. 1–14. [Google Scholar]
- Harsini, F.M.; Bui, A.A.; Rice, A.M.; Chebrolu, S.; Fuson, K.L.; Turtoi, A.; Bradberry, M.; Chapman, E.R.; Sutton, R.B. Structural Basis for the Distinct Membrane Binding Activity of the Homologous C2A Domains of Myoferlin and Dysferlin. J. Mol. Biol. 2019, 431, 2112–2126. [Google Scholar] [CrossRef] [PubMed]
- Therrien, C.; Fulvio, S.D.; Pickles, S.; Sinnreich, M. Characterization of lipid binding specificities of dysferlin C2 domains reveals novel interactions with phosphoinositides. Biochemistry 2009, 48, 2377–2384. [Google Scholar] [CrossRef] [PubMed]
- Marty, N.J.; Holman, C.L.; Abdullah, N.; Johnson, C.P. The C2 domains of otoferlin, dysferlin, and myoferlin alter the packing of lipid bilayers. Biochemistry 2013, 52, 5585–5592. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.P.; Chapman, E.R. Otoferlin is a calcium sensor that directly regulates SNARE-mediated membrane fusion. J. Cell Biol. 2010, 191, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Helfmann, S.; Neumann, P.; Tittmann, K.; Moser, T.; Ficner, R.; Reisinger, E. The Crystal Structure of the C2A Domain of Otoferlin Reveals an Unconventional Top Loop Region. J. Mol. Biol. 2011, 406, 479–490. [Google Scholar] [CrossRef]
- Harsini, F.M.; Chebrolu, S.; Fuson, K.L.; White, M.A.; Rice, A.M.; Sutton, R.B. FerA is a Membrane-Associating Four-Helix Bundle Domain in the Ferlin Family of Membrane-Fusion Proteins. Sci. Rep. 2018, 8, 10949. [Google Scholar] [CrossRef] [Green Version]
- De Morrée, A.; Hensbergen, P.J.; van Haagen, H.H.; Dragan, I.; Deelder, A.M.; AC’t Hoen, P.; Frants, R.R.; van der Maarel, S.M. Proteomic analysis of the dysferlin protein complex unveils its importance for sarcolemmal maintenance and integrity. PLoS ONE 2010, 5, e13854. [Google Scholar] [CrossRef]
- Doherty, K.R.; Demonbreun, A.R.; Wallace, G.Q.; Cave, A.; Posey, A.D.; Heretis, K.; Pytel, P.; McNally, E.M. The endocytic recycling protein EHD2 interacts with myoferlin to regulate myoblast fusion. J. Biol. Chem. 2008, 283, 20252–20260. [Google Scholar] [CrossRef]
- Bansal, D.; Miyake, K.; Vogel, S.S.; Groh, S.; Chen, C.-C.; Williamson, R.; McNeil, P.L.; Campbell, K.P. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature 2003, 423, 168–172. [Google Scholar] [CrossRef]
- Lukyanenko, V.; Muriel, J.M.; Bloch, R.J. Coupling of excitation to Ca2+ release is modulated by dysferlin. J. Physiol. 2017, 595, 5191–5207. [Google Scholar] [CrossRef] [PubMed]
- Lennon, N.J.; Kho, A.; Bacskai, B.J.; Perlmutter, S.L.; Hyman, B.T.; Brown, R.H. Dysferlin Interacts with Annexins A1 and A2 and Mediates Sarcolemmal Wound-healing. J. Biol. Chem. 2003, 278, 50466–50473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincent, A.E.; Rosa, H.S.; Alston, C.L.; Grady, J.P.; Rygiel, K.A.; Rocha, M.C.; Barresi, R.; Taylor, R.W.; Turnbull, D.M. Dysferlin mutations and mitochondrial dysfunction. Neuromuscul. Disord. 2016, 26, 782–788. [Google Scholar] [CrossRef] [Green Version]
- Pramono, Z.A.D.; Lai, P.S.; Tan, C.L.; Takeda, S.; Yee, W.C. Identification and characterization of a novel human dysferlin transcript: Dysferlin_v1. Hum. Genet. 2006, 120, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Lek, A.; Evesson, F.J.; Lemckert, F.A.; Redpath, G.M.I.; Lueders, A.-K.; Turnbull, L.; Whitchurch, C.B.; North, K.N.; Cooper, S.T. Calpains, cleaved mini-dysferlinC72, and L-type channels underpin calcium-dependent muscle membrane repair. J. Neurosci. 2013, 33, 5085–5094. [Google Scholar] [CrossRef]
- Redpath, G.M.I.; Woolger, N.; Piper, A.K.; Lemckert, F.A.; Lek, A.; Greer, P.A.; North, K.N.; Cooper, S.T. Calpain cleavage within dysferlin exon 40a releases a synaptotagmin-like module for membrane repair. Mol. Biol. Cell. 2014, 25, 3037–3048. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, V.; Lee, A.G. Synaptic vesicle fusion and synaptotagmin: 2B or not 2B? Nat. Neurosci. 2002, 5, 823–824. [Google Scholar] [CrossRef]
- Przybylski, R.J.; Szigeti, V.; Davidheiser, S.; Kirby, A.C. Calcium regulation of skeletal myogenesis. II. Extracellular and cell surface effects. Cell Calcium 1994, 15, 132–142. [Google Scholar] [CrossRef]
- Demonbreun, A.R.; Posey, A.D.; Heretis, K.; Swaggart, K.A.; Earley, J.U.; Pytel, P.; McNally, E.M. Myoferlin is required for insulin-like growth factor response and muscle growth. FASEB J. 2010, 24, 1284–1295. [Google Scholar] [CrossRef]
- Zhou, Y.; Xiong, L.; Zhang, Y.; Yu, R.; Jiang, X.; Xu, G. Quantitative proteomics identifies myoferlin as a novel regulator of A Disintegrin and Metalloproteinase 12 in HeLa cells. J. Proteom. 2016, 148, 94–104. [Google Scholar] [CrossRef]
- Galliano, M.F.; Huet, C.; Frygelius, J.; Polgren, A.; Wewer, U.M.; Engvall, E. Binding of ADAM12, a marker of skeletal muscle regeneration, to the muscle-specific actin-binding protein, α-actinin-2, is required for myoblast fusion. J. Biol. Chem. 2000, 275, 13933–13939. [Google Scholar] [CrossRef] [PubMed]
- Posey, A.D.; Pytel, P.; Gardikiotes, K.; Demonbreun, A.R.; Rainey, M.; George, M.; Band, H.; McNally, E.M. Endocytic recycling proteins EHD1 and EHD2 interact with fer-1-like-5 (Fer1L5) and mediate myoblast fusion. J. Biol. Chem. 2011, 286, 7379–7388. [Google Scholar] [CrossRef] [PubMed]
- Roux, I.; Safieddine, S.; Nouvian, R.; Grati, M.; Simmler, M.-C.; Bahloul, A.; Perfettini, I.; Le Gall, M.; Rostaing, P.; Hamard, G.; et al. Otoferlin, defective in a human deafness form, is essential for exocytosis at the auditory ribbon synapse. Cell 2006, 127, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.R.; Hanson, P.I.; An, S.; Jahn, R. Ca2+ regulates the interaction between synaptotagmin and syntaxin 1. J. Biol. Chem. 1995, 270, 23667–23671. [Google Scholar] [CrossRef] [PubMed]
- Mohrmann, R.; de Wit, H.; Connell, E.; Pinheiro, P.S.; Leese, C.; Bruns, D.; Davletov, B.; Verhage, M.; Sørensen, J.B. Synaptotagmin interaction with SNAP-25 governs vesicle docking, priming, and fusion triggering. J. Neurosci. 2013, 33, 14417–14430. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, N.A.; Drescher, M.J.; Drescher, D.G. Direct interaction of otoferlin with syntaxin 1A, SNAP-25, and the L-type voltage-gated calcium channel Cav1.3. J. Biol. Chem. 2009, 284, 1364–1372. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, N.A.; Drescher, M.J.; Morley, B.J.; Kelley, P.M.; Drescher, D.G. Calcium regulates molecular interactions of otoferlin with soluble NSF attachment protein receptor (SNARE) proteins required for hair cell exocytosis. J. Biol. Chem. 2014, 289, 8750–8766. [Google Scholar] [CrossRef]
- Hams, N.; Padmanarayana, M.; Qiu, W.; Johnson, C.P. Otoferlin is a multivalent calcium-sensitive scaffold linking SNAREs and calcium channels. Proc. Natl. Acad. Sci. USA 2017, 114, 8023–8028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Yu, C.; Leung, C.; Trane, A.; Lau, M.; Utokaparch, S.; Shaheen, F.; Sheibani, N.; Bernatchez, P. A new role for the muscle repair protein dysferlin in endothelial cell adhesion and angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 2196–2204. [Google Scholar] [CrossRef]
- Yu, C.; Sharma, A.; Trane, A.; Utokaparch, S.; Leung, C.; Bernatchez, P. Myoferlin gene silencing decreases Tie-2 expression in vitro and angiogenesis in vivo. Vascul. Pharmacol. 2011, 55, 26–33. [Google Scholar] [CrossRef]
- Bernatchez, P.N.; Sharma, A.; Kodaman, P.; Sessa, W.C. Myoferlin is critical for endocytosis in endothelial cells. Am. J. Physiol. Cell. Physiol. 2009, 297, C484–C492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, W.Q.; Xia, M.; Xu, M.; Boini, K.M.; Ritter, J.K.; Li, N.J.; Li, P.L. Lysosome fusion to the cell membrane is mediated by the dysferlin C2A domain in coronary arterial endothelial cells. J. Cell Sci. 2012, 125, 1225–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, C.; Shaheen, F.; Bernatchez, P.; Hackett, T.-L. Expression of myoferlin in human airway epithelium and its role in cell adhesion and zonula occludens-1 expression. PLoS ONE 2012, 7, e40478. [Google Scholar] [CrossRef] [PubMed]
- Stamer, W.D.; Hoffman, E.A.; Luther, J.M.; Hachey, D.L.; Schey, K.L. Protein profile of exosomes from trabecular meshwork cells. J. Proteom. 2011, 74, 796–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iacobuzio-Donahue, C.A.; Maitra, A.; Shen-Ong, G.L.; van Heek, T.; Ashfaq, R.; Meyer, R.; Walter, K.; Berg, K.; Hollingsworth, M.A.; Cameron, J.L.; et al. Discovery of Novel Tumor Markers of Pancreatic Cancer using Global Gene Expression Technology. Am. J. Pathol. 2002, 160, 1239–1249. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Bearss, D.J.; Browne, L.W.; Calaluce, R.; Nagle, R.B.; Von Hoff, D.D. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002, 62, 2890–2896. [Google Scholar] [PubMed]
- Amatschek, S.; Koenig, U.; Auer, H.; Steinlein, P.; Pacher, M.; Gruenfelder, A.; Dekan, G.; Vogl, S.; Kubista, E.; Heider, K.-H.; et al. Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res. 2004, 64, 844–856. [Google Scholar] [CrossRef]
- Yadav, A.; Kumar, B.; Lang, J.C.; Teknos, T.N.; Kumar, P. A muscle-specific protein “myoferlin” modulates IL-6/STAT3 signaling by chaperoning activated STAT3 to nucleus. Oncogene 2017, 36, 6374–6382. [Google Scholar] [CrossRef]
- McKinney, K.Q.; Lee, Y.Y.; Choi, H.S.; Groseclose, G.; Iannitti, D.A.; Martinie, J.B.; Russo, M.W.; Lundgren, D.H.; Han, D.K.; Bonkovsky, H.L.; et al. Discovery of putative pancreatic cancer biomarkers using subcellular proteomics. J. Proteom. 2011, 74, 79–88. [Google Scholar] [CrossRef]
- Turtoi, A.; Musmeci, D.; Wang, Y.; Dumont, B.; Somja, J.; Bevilacqua, G.; De Pauw, E.; Delvenne, P.; Castronovo, V. Identification of novel accessible proteins bearing diagnostic and therapeutic potential in human pancreatic ductal adenocarcinoma. J. Proteome Res. 2011, 10, 4302–4313. [Google Scholar] [CrossRef]
- McKinney, K.Q.; Lee, J.-G.; Sindram, D.; Russo, M.W.; Han, D.K.; Bonkovsky, H.L.; Hwang, S.-I. Identification of differentially expressed proteins from primary versus metastatic pancreatic cancer cells using subcellular proteomics. Cancer Genom. Proteom. 2012, 9, 257–263. [Google Scholar]
- Wang, W.S.; Liu, X.H.; Liu, L.X.; Lou, W.H.; Jin, D.Y.; Yang, P.Y.; Wang, X.L. ITRAQ-based quantitative proteomics reveals myoferlin as a novel prognostic predictor in pancreatic adenocarcinoma. J. Proteom. 2013, 91, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Adam, P.J.; Boyd, R.; Tyson, K.L.; Fletcher, G.C.; Stamps, A.; Hudson, L.; Poyser, H.R.; Redpath, N.; Griffiths, M.; Steers, G.; et al. Comprehensive proteomic analysis of breast cancer cell membranes reveals unique proteins with potential roles in clinical cancer. J. Biol. Chem. 2003, 278, 6482–6489. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.; Yu, C.; Lin, M.I.; Tognon, C.; Bernatchez, P. Expression of myoferlin in human and murine carcinoma tumors: Role in membrane repair, cell proliferation, and tumorigenesis. Am. J. Pathol. 2013, 182, 1900–1909. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, C.; Hampl, V.; Holzer, K.; Aigner, A.; Penkava, J.; Frank, N.; Martin, D.E.; Maier, K.C.; Waldburger, N.; Roessler, S.; et al. The novel MKL target gene myoferlin modulates expansion and senescence of hepatocellular carcinoma. Oncogene 2017, 36, 3464–3476. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Brown, N.V.; Swanson, B.J.; Schmitt, A.C.; Old, M.; Ozer, E.; Agrawal, A.; Schuller, D.E.; Teknos, T.N.; Kumar, P. High expression of myoferlin is associated with poor outcome in oropharyngeal squamous cell carcinoma patients and is inversely associated with HPV-status. Oncotarget 2016, 7, 18665–18677. [Google Scholar] [CrossRef] [PubMed]
- Song, D.H.; Ko, G.H.; Lee, J.H.; Lee, J.S.; Yang, J.W.; Kim, M.H.; An, H.J.; Kang, M.H.; Jeon, K.N.; Kim, D.C. Prognostic role of myoferlin expression in patients with clear cell renal cell carcinoma. Oncotarget 2017, 8, 89033–89039. [Google Scholar] [CrossRef] [Green Version]
- Koh, H.M.; An, H.J.; Ko, G.H.; Lee, J.H.; Lee, J.S.; Kim, D.C.; Seo, D.H.; Song, D.H. Identification of Myoferlin Expression for Prediction of Subsequent Primary Malignancy in Patients With Clear Cell Renal Cell Carcinoma. In Vivo 2019, 33, 1103–1108. [Google Scholar] [CrossRef]
- Kim, M.H.; Song, D.H.; Ko, G.H.; Lee, J.H.; Kim, D.C.; Yang, J.W.; Lee, H.I.; An, H.J.; Lee, J.S. Myoferlin expression and its correlation with FIGO histologic grading in early-stage endometrioid carcinoma. J. Pathol. Transl. Med. 2018, 52, 93–97. [Google Scholar] [CrossRef]
- Welton, J.L.; Khanna, S.; Giles, P.J.; Brennan, P.; Brewis, I.A.; Staffurth, J.; Mason, M.D.; Clayton, A. Proteomics analysis of bladder cancer exosomes. Mol. Cell. Proteom. 2010, 9, 1324–1338. [Google Scholar] [CrossRef]
- Mathivanan, S.; Lim, J.W.E.; Tauro, B.J.; Ji, H.; Moritz, R.L.; Simpson, R.J. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell. Proteom. 2010, 9, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Beckler, M.D.; Higginbotham, J.N.; Franklin, J.L.; Ham, A.-J.; Halvey, P.J.; Imasuen, I.E.; Whitwell, C.; Li, M.; Liebler, D.C.; Coffey, R.J. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol. Cell. Proteom. 2013, 12, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Greening, D.W.; Barnes, T.W.; Lim, J.W.; Tauro, B.J.; Rai, A.; Xu, R.; Adda, C.; Mathivanan, S.; Zhao, W.; et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics 2013, 13, 1672–1686. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.-S.; Choi, D.-Y.; Hong, B.S.; Jang, S.C.; Kim, D.-K.; Lee, J.; Kim, Y.-K.; Kim, K.P.; Gho, Y.S. Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. J. Extracell. Vesicles 2012, 1, 18704. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Peng, P.; Chen, S.; Li, L.; Zhang, M.; Cao, D.; Yang, J.; Li, H.; Gui, T.; Li, X.; et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J. Proteom. 2013, 80, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Sandvig, K.; Llorente, A. Proteomic analysis of microvesicles released by the human prostate cancer cell line PC-3. Mol. Cell. Proteom. 2012, 11, M111-012914. [Google Scholar] [CrossRef]
- Blomme, A.; Fahmy, K.; Peulen, O.J.; Costanza, B.; Fontaine, M.; Struman, I.; Baiwir, D.; De Pauw, E.; Thiry, M.; Bellahcène, A.; et al. Myoferlin is a novel exosomal protein and functional regulator of cancer-derived exosomes. Oncotarget 2016, 7, 83669–83683. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef] [Green Version]
- Anaya, J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comp. Sci. 2016, 2, e67. [Google Scholar] [CrossRef]
- Tang, H.; Wei, P.; Chang, P.; Li, Y.; Yan, D.; Liu, C.; Hassan, M.; Li, D. Genetic polymorphisms associated with pancreatic cancer survival: A genome-wide association study. Int. J. Cancer 2017, 141, 678–686. [Google Scholar] [CrossRef]
- Ding, F.; Tang, H.; Nie, D.; Xia, L. Long non-coding RNA Fer-1-like family member 4 is overexpressed in human glioblastoma and regulates the tumorigenicity of glioma cells. Oncol. Lett. 2017, 14, 2379–2384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, L.; Nie, D.; Wang, G.; Sun, C.; Chen, G. FER1L4/miR-372/E2F1 works as a ceRNA system to regulate the proliferation and cell cycle of glioma cells. J. Cell. Mol. Med. 2019, 23, 3224–3233. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Ge, A.; Pang, D.; Zhao, Y.; Xu, S. Long noncoding RNA FER1L4 acts as an oncogenic driver in human pan-cancer. J. Cell. Physiol. 2019, 1859, 46. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-X.; Chen, C.-P.; Zhang, N.; Wang, T.-X. Low-expression of lncRNA FER1L4 might be a prognostic marker in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2310–2314. [Google Scholar]
- Song, H.; Sun, W.; Ye, G.; Ding, X.; Liu, Z.; Zhang, S.; Xia, T.; Xiao, B.; Xi, Y.; Guo, J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J. Transl. Med. 2013, 11, 225. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Ren, Z. Overexpression of LncRNA FER1L4 in endometrial carcinoma is associated with favorable survival outcome. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8113–8118. [Google Scholar]
- Eisenberg, M.C.; Kim, Y.; Li, R.; Ackerman, W.E.; Kniss, D.A.; Friedman, A. Mechanistic modeling of the effects of myoferlin on tumor cell invasion. Proc. Natl. Acad. Sci. USA 2011, 108, 20078–20083. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Ackerman, W.E.; Mihai, C.; Volakis, L.I.; Ghadiali, S.; Kniss, D.A. Myoferlin depletion in breast cancer cells promotes mesenchymal to epithelial shape change and stalls invasion. PLoS ONE 2012, 7, e39766. [Google Scholar] [CrossRef]
- Volakis, L.I.; Li, R.; Ackerman, W.E.; Mihai, C.; Bechel, M.; Summerfield, T.L.; Ahn, C.S.; Powell, H.M.; Zielinski, R.; Rosol, T.J.; et al. Loss of myoferlin redirects breast cancer cell motility towards collective migration. PLoS ONE 2014, 9, e86110. [Google Scholar] [CrossRef]
- Blackstone, B.N.; Li, R.; Ackerman, W.E.; Ghadiali, S.N.; Powell, H.M.; Kniss, D.A. Myoferlin depletion elevates focal adhesion kinase and paxillin phosphorylation and enhances cell-matrix adhesion in breast cancer cells. Am. J. Physiol. Cell. Physiol. 2015, 308, C642–C649. [Google Scholar] [CrossRef] [Green Version]
- Barnhouse, V.R.; Weist, J.L.; Shukla, V.C.; Ghadiali, S.N.; Kniss, D.A.; Leight, J.L. Myoferlin regulates epithelial cancer cell plasticity and migration through autocrine TGF-β1 signaling. Oncotarget 2018, 9, 19209–19222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhou, P.; Meng, A.; Zhang, R.; Zhou, Y. Down-regulating Myoferlin inhibits the vasculogenic mimicry of melanoma via decreasing MMP-2 and inducing mesenchymal-to-epithelial transition. J. Cell. Mol. Med. 2017, 155, 739. [Google Scholar] [CrossRef] [PubMed]
- Turtoi, A.; Blomme, A.; Bellahcène, A.; Gilles, C.; Hennequière, V.; Peixoto, P.; Bianchi, E.; Noël, A.; De Pauw, E.; Lifrange, E.; et al. Myoferlin is a key regulator of EGFR activity in breast cancer. Cancer Res. 2013, 73, 5438–5448. [Google Scholar] [CrossRef] [PubMed]
- Nylandsted, J.; Becker, A.C.; Bunkenborg, J.; Andersen, J.S.; Dengjel, J.; Jäättelä, M. ErbB2-associated changes in the lysosomal proteome. Proteomics 2011, 11, 2830–2838. [Google Scholar] [CrossRef] [PubMed]
- Örtegren, U.; Aboulaich, N.; Öst, A.; Strålfors, P. A new role for caveolae as metabolic platforms. Trends Endocrinol. Metab. 2007, 18, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Blomme, A.; Costanza, B.; de Tullio, P.; Thiry, M.; Van Simaeys, G.; Boutry, S.; Doumont, G.; Di Valentin, E.; Hirano, T.; Yokobori, T.; et al. Myoferlin regulates cellular lipid metabolism and promotes metastases in triple-negative breast cancer. Oncogene 2017, 36, 2116–2130. [Google Scholar] [CrossRef] [PubMed]
- Piper, A.-K.; Ross, S.E.; Redpath, G.M.; Lemckert, F.A.; Woolger, N.; Bournazos, A.; Greer, P.A.; Sutton, R.B.; Cooper, S.T. Enzymatic cleavage of myoferlin releases a dual C2-domain module linked to ERK signalling. Cell. Signal. 2017, 33, 30–40. [Google Scholar] [CrossRef]
- Fahmy, K.; Gonzalez, A.; Arafa, M.; Peixoto, P.; Bellahcène, A.; Turtoi, A.; Delvenne, P.; Thiry, M.; Castronovo, V.; Peulen, O.J. Myoferlin plays a key role in VEGFA secretion and impacts tumor-associated angiogenesis in human pancreas cancer. Int. J. Cancer 2016, 138, 652–663. [Google Scholar] [CrossRef]
- Rademaker, G.; Hennequière, V.; Brohée, L.; Nokin, M.-J.; Lovinfosse, P.; Durieux, F.; Gofflot, S.; Bellier, J.; Costanza, B.; Herfs, M.; et al. Myoferlin controls mitochondrial structure and activity in pancreatic ductal adenocarcinoma, and affects tumor aggressiveness. Oncogene 2018, 66, 1–15. [Google Scholar] [CrossRef]
- Rademaker, G.; Costanza, B.; Bellier, J.; Herfs, M.; Peiffer, R.; Agirman, F.; Maloujahmoum, N.; Habraken, Y.; Delvenne, P.; Bellahcène, A.; et al. Human colon cancer cells highly express myoferlin to maintain a fit mitochondrial network and escape p53-driven apoptosis. Oncogenesis 2019, 8, 21. [Google Scholar] [CrossRef]
- LeBleu, V.S.; O’Connell, J.T.; Gonzalez Herrera, K.N.; Wikman, H.; Pantel, K.; Haigis, M.C.; de Carvalho, F.M.; Damascena, A.; Domingos Chinen, L.T.; Rocha, R.M.; et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014, 16, 992–1003. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E.; Payen, V.L.; Pérez-Escuredo, J.; De Saedeleer, C.J.; Danhier, P.; Copetti, T.; Dhup, S.; Tardy, M.; Vazeille, T.; Bouzin, C.; et al. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014, 8, 754–766. [Google Scholar] [CrossRef] [PubMed]
- Rademaker, G.; Costanza, B.; Anania, S.; Agirman, F.; Maloujahmoum, N.; Di Valentin, E.; Goval, J.J.; Bellahcène, A.; Castronovo, V.; Peulen, O.J. Myoferlin Contributes to the Metastatic Phenotype of Pancreatic Cancer Cells by Enhancing Their Migratory Capacity through the Control of Oxidative Phosphorylation. Cancers 2019, 11, 853. [Google Scholar] [CrossRef] [PubMed]
- Majumder, S.; Raimondo, M.; Taylor, W.R.; Yab, T.C.; Berger, C.K.; Dukek, B.A.; Cao, X.; Foote, P.H.; Wu, C.W.; Devens, M.E.; et al. Methylated DNA in Pancreatic Juice Distinguishes Patients with Pancreatic Cancer from Controls. Clin. Gastroenterol. Hepatol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Song, D.H.; Ko, G.H.; Lee, J.H.; Lee, J.S.; Lee, G.-W.; Kim, H.C.; Yang, J.W.; Heo, R.W.; Roh, G.S.; Han, S.-Y.; et al. Myoferlin expression in non-small cell lung cancer: Prognostic role and correlation with VEGFR-2 expression. Oncol. Lett. 2016, 11, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, N.; Wu, S.; Cui, H.; An, X.; Yang, Y. Long non-coding RNA FER1L4 inhibits cell proliferation and metastasis through regulation of the PI3K/AKT signaling pathway in lung cancer cells. Mol. Med. Rep. 2019, 20, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhang, L.; Guo, A.; Liu, L.C.; Yu, F.; Diao, N.; Xu, C.; Wang, D. Overexpression of FER1L4 promotes the apoptosis and suppresses epithelial-mesenchymal transition and stemness markers via activating PI3K/AKT signaling pathway in osteosarcoma cells. Pathol. Res. Pract. 2019, 215, 152412. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhang, C.-Q.; Li, H.-L.; Gu, J.; Miao, G.-Y.; Cai, H.-Y.; Wang, J.-K.; Zhang, L.-J.; Song, Y.-M.; Tian, Y.-H.; et al. LncRNA FER1L4 suppressed cancer cell growth and invasion in esophageal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2638–2645. [Google Scholar] [PubMed]
- Wang, X.; Dong, K.; Jin, Q.; Ma, Y.; Yin, S.; Wang, S. Upregulation of lncRNA FER1L4 suppresses the proliferation and migration of the hepatocellular carcinoma via regulating PI3K/AKT signal pathway. J. Cell. Biochem. 2019, 120, 6781–6788. [Google Scholar] [CrossRef]
- Liu, Z.; Shao, Y.; Tan, L.; Shi, H.; Chen, S.; Guo, J. Clinical significance of the low expression of FER1L4 in gastric cancer patients. Tumour Biol. 2014, 35, 9613–9617. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Chen, S.; Jiang, Z.; Shao, Y.; Jiang, X.; Li, P.; Xiao, B.; Guo, J. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci. Rep. 2015, 5, 13445. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Liao, Q.; Jiang, X.; Shao, Y.; Xiao, B.; Xi, Y.; Guo, J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci. Rep. 2014, 4, 6088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Sun, B.; Liu, C.; Zhao, S.; Zhang, D.; Yu, F.; Yan, D. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 2015, 106, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Huang, J.; Wang, W.; Xu, J.; Yin, M.; Cheng, N.; Yin, J. Long non-coding RNA Fer-1-like protein 4 acts as a tumor suppressor via miR-106a-5p and predicts good prognosis in hepatocellular carcinoma. Cancer Biomark. 2017, 20, 55–65. [Google Scholar] [CrossRef]
- Fei, D.; Zhang, X.; Liu, J.; Tan, L.; Xing, J.; Zhao, D.; Zhang, Y. Long Noncoding RNA FER1L4 Suppresses Tumorigenesis by Regulating the Expression of PTEN Targeting miR-18a-5p in Osteosarcoma. Cell. Physiol. Biochem. 2018, 51, 1364–1375. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zou, B.; Tian, T.; Luo, X.; Mao, B.; Zhang, X.; Lei, H. Overexpression of the lncRNA FER1L4 inhibits paclitaxel tolerance of ovarian cancer cells via the regulation of the MAPK signaling pathway. J. Cell. Biochem. 2018, 120, 7581–7589. [Google Scholar] [CrossRef]
- Théry, C. Cancer: Diagnosis by extracellular vesicles. Nature 2015, 523, 161–162. [Google Scholar] [CrossRef]
- Ciardiello, C.; Cavallini, L.; Spinelli, C.; Yang, J.; Reis-Sobreiro, M.; de Candia, P.; Minciacchi, V.R.; Di Vizio, D. Focus on Extracellular Vesicles: New Frontiers of Cell-to-Cell Communication in Cancer. Int. J. Mol. Sci. 2016, 17, 175. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Anderson, R.G.; Ghiraldeli, L.P.; Pardee, T.S. Mitochondria in cancer metabolism, an organelle whose time has come? Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Jingjie, L.; He, Y.; Yang, F.; Hao, Y.; Jin, W.; Wu, J.; Sun, Z.; Li, Y.; Chen, Y.; et al. A small molecule targeting myoferlin exerts promising anti-tumor effects on breast cancer. Nat. Commun. 2018, 9, 3726. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, Y.; Shao, T.; Pei, H.; Guo, W.; Mi, D.; Krimm, I.; Zhang, Y.; Wang, P.; Wang, X.; et al. Modification and Biological Evaluation of a Series of 1,5-Diaryl-1,2,4-triazole Compounds as Novel Agents against Pancreatic Cancer Metastasis through Targeting Myoferlin. J. Med. Chem. 2019, 62, 4949–4966. [Google Scholar] [CrossRef] [PubMed]
- Sondergaard, P.C.; Griffin, D.A.; Pozsgai, E.R.; Johnson, R.W.; Grose, W.E.; Heller, K.N.; Shontz, K.M.; Montgomery, C.L.; Liu, J.; Clark, K.R.; et al. AAV.Dysferlin Overlap Vectors Restore Function in Dysferlinopathy Animal Models. Ann. Clin. Transl. Neurol. 2015, 2, 256–270. [Google Scholar] [CrossRef] [PubMed]
- Escobar, H.; Schöwel, V.; Spuler, S.; Marg, A.; Izsvák, Z. Full-length Dysferlin Transfer by the Hyperactive Sleeping Beauty Transposase Restores Dysferlin-deficient Muscle. Mol. Ther. Nucleic Acids 2016, 5, e277. [Google Scholar] [CrossRef] [PubMed]
- Potter, R.A.; Griffin, D.A.; Sondergaard, P.C.; Johnson, R.W.; Pozsgai, E.R.; Heller, K.N.; Peterson, E.L.; Lehtimäki, K.K.; Windish, H.P.; Mittal, P.J.; et al. Systemic Delivery of Dysferlin Overlap Vectors Provides Long-Term Gene Expression and Functional Improvement for Dysferlinopathy. Hum. Gene Ther. 2017. hum.2017.062. [Google Scholar] [CrossRef]
- Llanga, T.; Nagy, N.; Conatser, L.; Dial, C.; Sutton, R.B.; Hirsch, M.L. Structure-Based Designed Nano-Dysferlin Significantly Improves Dysferlinopathy in BLA/J Mice. Mol. Ther. 2017, 25, 2150–2162. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Kim, Y.; Yoon, Y.-E.; Kim, Y.-J.; Oh, S.-G.; Jang, J.-H.; Kim, E. Development of efficient adeno-associated virus (AAV)-mediated gene delivery system with a phytoactive material for targeting human melanoma cells. New Biotechnol. 2017, 37, 194–199. [Google Scholar] [CrossRef]
- Chow, R.D.; Guzman, C.D.; Wang, G.; Schmidt, F.; Youngblood, M.W.; Ye, L.; Errami, Y.; Dong, M.B.; Martinez, M.A.; Zhang, S.; et al. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat. Neurosci. 2017, 20, 1329–1341. [Google Scholar] [CrossRef] [Green Version]
- Hodge, R.; Narayanavari, S.A.; Izsvák, Z.; Ivics, Z. Wide Awake and Ready to Move: 20 Years of Non-Viral Therapeutic Genome Engineering with the Sleeping Beauty Transposon System. Hum. Gene Ther. 2017, 28, 842–855. [Google Scholar] [CrossRef]



| Protein Name (Uniprot Number) | Gene Name | Chromosome Mapping | Main Protein Size |
|---|---|---|---|
| Sperm vesicle fusion protein FER-1 (Q17388) | fer-1 | 2034 AA (235 KDa) | |
| Dysferlin (O75923) | Fer1-Like 1 Fer1L1 | 2p13.2 | 2080 AA (237 KDa) |
| Otoferlin (Q9HC10) | Fer1-Like Fer1L2 | 2p23.3 | 1997 AA (227 KDa) |
| Myoferlin (Q9NZM1) | Fer1-Like 3 Fer1L3 | 10q23.33 | 2061 AA (230 KDa) |
| FER1L4 (A9Z1Z3) | Fer1-Like 4 Fer1L4 | 20q11.22 | pseudogene |
| FER1L5 (A0AVI2) | Fer1-Like 5 Fer1L5 | 2q11.2 | 2057 AA (238 KDa) |
| FER1L6 (Q2WGJ9) | Fer1-Like 6 Fer1L6 | 8q24.13 | 1857 AA (209 KDa) |
| Gene Name | Gene Length | Number of Exons | Transcript Size | Number of Variants |
|---|---|---|---|---|
| Fer-1 | 8.6 kb | 21 | 6.2 kb | 3 |
| Fer1-Like 1 Fer1L1 (DYSF) | 233 kb | 55 | 0.5–6.7 kb | 19 |
| Fer1-Like 2 Fer1L2 (OTOF) | 121 kb | 47 | 0.5–7.2 kb | 7 |
| Fer1-Like 3 Fer1L3 (MYOF) | 180 kb | 54 | 0.4–6.7 kb | 9 |
| Fer1-Like 4 Fer1L4 | 48 kb | 43 | 0.2–5.9 kb | 13 |
| Fer1-Like 5 Fer1L5 | 64 kb | 53 | 3.5–6.5 kb | 7 |
| Fer1-Like 6 Fer1L6 | 278 kb | 41 | 6 kb | 1 |
| Positive Association | Negative Association | ||||
|---|---|---|---|---|---|
| Cohort | Cox Coefficient | p-Value | Cohort | Cox Coefficient | p-Value |
| DYSF EXPRESSION | |||||
| CESC | 0.266 | 4.20e−02 | SARC | −0.277 | 1.00e−02 |
| STAD | 0.171 | 4.80e−02 | KIRC | −0.220 | 1.00e−02 |
| OTOF EXPRESSION | |||||
| KIRC | 0.377 | 1.50e−06 | BLCA | −0.275 | 4.50e−04 |
| KIRP | 0.413 | 4.90e−03 | SKCM | −0.169 | 1.40e−02 |
| MYOF EXPRESSION | |||||
| LGG | 0.441 | 1.40e−05 | SKCM | −0.163 | 1.90e−02 |
| PAAD | 0.561 | 1.70e−05 | |||
| LAML | 0.215 | 4.70e−02 | |||
| FER1L4 EXPRESSION | |||||
| KIRC | 0.356 | 5.20e−06 | BLCA | −0.383 | 2.90e−06 |
| KIRP | 0.492 | 1.10e−03 | SKCM | −0.225 | 1.10e−03 |
| LGG | 0.244 | 4.00e−03 | |||
| FER1L5 EXPRESSION | |||||
| LUAD | −0.199 | 1.30e−02 | |||
| FER1L6 EXPRESSION | |||||
| KIRC | −0.160 | 4.80e−02 | |||
| READ | −0.401 | 4.90e−02 | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peulen, O.; Rademaker, G.; Anania, S.; Turtoi, A.; Bellahcène, A.; Castronovo, V. Ferlin Overview: From Membrane to Cancer Biology. Cells 2019, 8, 954. https://doi.org/10.3390/cells8090954
Peulen O, Rademaker G, Anania S, Turtoi A, Bellahcène A, Castronovo V. Ferlin Overview: From Membrane to Cancer Biology. Cells. 2019; 8(9):954. https://doi.org/10.3390/cells8090954
Chicago/Turabian StylePeulen, Olivier, Gilles Rademaker, Sandy Anania, Andrei Turtoi, Akeila Bellahcène, and Vincent Castronovo. 2019. "Ferlin Overview: From Membrane to Cancer Biology" Cells 8, no. 9: 954. https://doi.org/10.3390/cells8090954
APA StylePeulen, O., Rademaker, G., Anania, S., Turtoi, A., Bellahcène, A., & Castronovo, V. (2019). Ferlin Overview: From Membrane to Cancer Biology. Cells, 8(9), 954. https://doi.org/10.3390/cells8090954