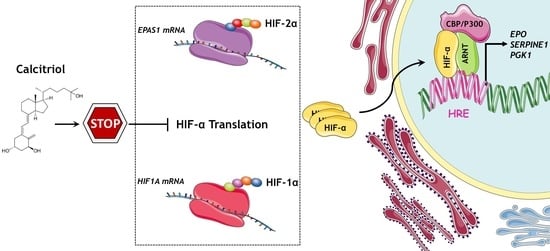
Calcitriol Suppresses HIF-1 and HIF-2 Transcriptional Activity by Reducing HIF-1/2α Protein Levels via a VDR-Independent Mechanism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture, Transfection, and Luciferase Assays
2.2. siRNA-Mediated Silencing
2.3. SDS-PAGE and Western Blot
2.4. Polysome Profiling
2.5. RNA Extraction, cDNA Preparation, and Quantitative PCR
2.6. Cell Proliferation Assay
2.7. Statistical Analysis
3. Results
3.1. Calcitriol Reduces Both HIF-1α and HIF-2α Protein Levels and Hampers HIF-1 and HIF-2 Transcriptional Activity in Dose and Time Dependent Manner
3.2. Negative Regulation of HIF-1/2α Protein Levels and HIFs Transcriptional Activity by Calcitriol is not Mediated by VDR Activation by Calcitriol
3.3. Calcitriol Suppresses HIF1A and EPAS1 mRNA Translation by Downregulating Components of PI3K/Akt/mTOR Signaling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Haidari, F.; Abiri, B.; Iravani, M.; Razavi, S.M.; Vafa, M. The Effects of UVB and Vitamin D on Decreasing Risk of Colorectal Cancer Incidence and Mortality: A Review of the Epidemiology, Clinical Trials, and Mechanisms. Nutr Cancer 2019, 71, 709–717. [Google Scholar] [CrossRef]
- Deeb, K.K.; Trump, D.L.; Johnson, C.S. Vitamin D signalling pathways in cancer: Potential for anticancer therapeutics. Nat. Rev. Cancer 2007, 7, 684–700. [Google Scholar] [CrossRef]
- Feldman, D.; Krishnan, A.V.; Swami, S.; Giovannucci, E.; Feldman, B.J. The role of vitamin D in reducing cancer risk and progression. Nat. Rev. Cancer 2014, 14, 342–357. [Google Scholar] [CrossRef]
- Grant, W.B.; Boucher, B.J. Randomized controlled trials of vitamin D and cancer incidence: A modeling study. PLoS ONE 2017, 12, e0176448. [Google Scholar] [CrossRef] [Green Version]
- Minisola, S.; Ferrone, F.; Danese, V.; Cecchetti, V.; Pepe, J.; Cipriani, C.; Colangelo, L. Controversies Surrounding Vitamin D: Focus on Supplementation and Cancer. Int. J. Environ. Res. Public Health 2019, 16, 189. [Google Scholar] [CrossRef] [Green Version]
- Pilz, S.; Zittermann, A.; Trummer, C.; Theiler-Schwetz, V.; Lerchbaum, E.; Keppel, M.H.; Grubler, M.R.; Marz, W.; Pandis, M. Vitamin D testing and treatment: A narrative review of current evidence. Endocr Connect. 2019, 8, R27–R43. [Google Scholar] [CrossRef] [Green Version]
- Jeon, S.M.; Shin, E.A. Exploring vitamin D metabolism and function in cancer. Exp. Mol. Med. 2018, 50, 20. [Google Scholar] [CrossRef] [Green Version]
- Haussler, M.R.; Jurutka, P.W.; Mizwicki, M.; Norman, A.W. Vitamin D receptor (VDR)-mediated actions of 1alpha,25(OH)(2)vitamin D(3): Genomic and non-genomic mechanisms. Best Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 543–559. [Google Scholar] [CrossRef]
- Hii, C.S.; Ferrante, A. The Non-Genomic Actions of Vitamin, D. Nutrients 2016, 8, 135. [Google Scholar] [CrossRef] [Green Version]
- Moeenrezakhanlou, A.; Nandan, D.; Reiner, N.E. Identification of a Calcitriol-Regulated Sp-1 Site in the Promoter of Human CD14 using a Combined Western Blotting Electrophoresis Mobility Shift Assay (WEMSA). Biol. Proced. Online 2008, 10, 29–35. [Google Scholar] [CrossRef] [Green Version]
- Moeenrezakhanlou, A.; Shephard, L.; Lam, L.; Reiner, N.E. Myeloid cell differentiation in response to calcitriol for expressionCD11bandCD14is regulated by myeloid zinc finger-1 protein downstream of phosphatidylinositol 3-kinase. J. Leukoc. Biol. 2008, 84, 519–528. [Google Scholar] [CrossRef] [Green Version]
- Khanal, R.C.; Nemere, I. The ERp57/GRp58/1,25D3-MARRS receptor: Multiple functional roles in diverse cell systems. Curr. Med. Chem. 2007, 14, 1087–1093. [Google Scholar] [CrossRef]
- Datta Mitra, A.; Raychaudhuri, S.P.; Abria, C.J.; Mitra, A.; Wright, R.; Ray, R.; Kundu-Raychaudhuri, S. 1alpha,25-Dihydroxyvitamin-D3-3-bromoacetate regulates AKT/mTOR signaling cascades: A therapeutic agent for psoriasis. J. Investig. Dermatol. 2013, 133, 1556–1564. [Google Scholar] [CrossRef] [Green Version]
- Kemmochi, S.; Fujimoto, H.; Woo, G.H.; Hirose, M.; Nishikawa, A.; Mitsumori, K.; Shibutani, M. Preventive effects of calcitriol on the development of capsular invasive carcinomas in a rat two-stage thyroid carcinogenesis model. J. Vet. Med. Sci 2011, 73, 655–664. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Johnson, C.S.; Trump, D.L. Mechanistic Insights of Vitamin D Anticancer Effects. Vitam. Horm. 2016, 100, 395–431. [Google Scholar] [CrossRef]
- Liu, P.; Cheng, H.; Roberts, T.M.; Zhao, J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009, 8, 627–644. [Google Scholar] [CrossRef] [Green Version]
- Patra, K.; Jana, S.; Sarkar, A.; Mandal, D.P.; Bhattacharjee, S. The inhibition of hypoxia-induced angiogenesis and metastasis by cinnamaldehyde is mediated by decreasing HIF-1alpha protein synthesis via PI3K/Akt pathway. Biofactors 2019, 45, 401–415. [Google Scholar] [CrossRef]
- Soler, A.; Angulo-Urarte, A.; Graupera, M. PI3K at the crossroads of tumor angiogenesis signaling pathways. Mol. Cell. Oncol. 2015, 2, e975624. [Google Scholar] [CrossRef] [Green Version]
- Schito, L.; Semenza, G.L. Hypoxia-Inducible Factors: Master Regulators of Cancer Progression. Trends Cancer 2016, 2, 758–770. [Google Scholar] [CrossRef] [Green Version]
- Ivan, M.; Kaelin, W.G. The EGLN-HIF O 2 -Sensing System: Multiple Inputs and Feedbacks. Mol. Cell 2017, 66, 772–779. [Google Scholar] [CrossRef] [Green Version]
- Dery, M.A.; Michaud, M.D.; Richard, D.E. Hypoxia-inducible factor 1: Regulation by hypoxic and non-hypoxic activators. Int. J. Biochem. Cell Biol. 2005, 37, 535–540. [Google Scholar] [CrossRef]
- Koh, M.Y.; Powis, G. Passing the baton: The HIF switch. Trends Biochem. Sci. 2012, 37, 364–372. [Google Scholar] [CrossRef] [Green Version]
- Agani, F.; Jiang, B.H. Oxygen-independent regulation of HIF-1: Novel involvement of PI3K/AKT/mTOR pathway in cancer. Curr. Cancer Drug Targets 2013, 13, 245–251. [Google Scholar] [CrossRef]
- Befani, C.; Liakos, P. The role of hypoxia-inducible factor-2 alpha in angiogenesis. J. Cell. Physiol. 2018, 233, 9087–9098. [Google Scholar] [CrossRef]
- Fukuda, R.; Hirota, K.; Fan, F.; Jung, Y.D.; Ellis, L.M.; Semenza, G.L. Insulin-like growth factor 1 induces hypoxia-inducible factor 1-mediated vascular endothelial growth factor expression, which is dependent on MAP kinase and phosphatidylinositol 3-kinase signaling in colon cancer cells. J. Biol. Chem. 2002, 277, 38205–38211. [Google Scholar] [CrossRef] [Green Version]
- Masoud, G.N.; Li, W. HIF-1alpha pathway: Role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 2015, 5, 378–389. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Qi, Y.; Zhang, Y.; Chen, J.; Wu, T.; Gu, Y. Regulation of angiogenic factors by the PI3K/Akt pathway in A549 lung cancer cells under hypoxic conditions. Oncol. Lett. 2017, 13, 2909–2914. [Google Scholar] [CrossRef] [Green Version]
- Flügel, D.; Görlach, A.; Michiels, C.; Kietzmann, T. Glycogen Synthase Kinase 3 Phosphorylates Hypoxia-Inducible Factor 1α and Mediates Its Destabilization in a VHL-Independent Manner. Mol. Cell. Biol. 2007, 27, 3253–3265. [Google Scholar] [CrossRef] [Green Version]
- Kietzmann, T.; Mennerich, D.; Dimova, E.Y. Hypoxia-Inducible Factors (HIFs) and Phosphorylation: Impact on Stability, Localization, and Transactivity. Front. Cell Dev. Biol. 2016, 4, 11. [Google Scholar] [CrossRef]
- Gkotinakou, I.M.; Befani, C.; Simos, G.; Liakos, P. ERK1/2 phosphorylates HIF-2alpha and regulates its activity by controlling its CRM1-dependent nuclear shuttling. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [Green Version]
- Mylonis, I.; Chachami, G.; Paraskeva, E.; Simos, G. Atypical CRM1-dependent nuclear export signal mediates regulation of hypoxia-inducible factor-1alpha by MAPK. J. Biol. Chem. 2008, 283, 27620–27627. [Google Scholar] [CrossRef] [Green Version]
- Mylonis, I.; Chachami, G.; Samiotaki, M.; Panayotou, G.; Paraskeva, E.; Kalousi, A.; Georgatsou, E.; Bonanou, S.; Simos, G. Identification of MAPK phosphorylation sites and their role in the localization and activity of hypoxia-inducible factor-1alpha. J. Biol. Chem. 2006, 281, 33095–33106. [Google Scholar] [CrossRef] [Green Version]
- Loboda, A.; Jozkowicz, A.; Dulak, J. HIF-1 and HIF-2 transcription factors--similar but not identical. Mol. Cells 2010, 29, 435–442. [Google Scholar] [CrossRef]
- Majmundar, A.J.; Wong, W.J.; Simon, M.C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 2010, 40, 294–309. [Google Scholar] [CrossRef] [Green Version]
- Keith, B.; Johnson, R.S.; Simon, M.C. HIF1alpha and HIF2alpha: Sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer 2011, 12, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Tolonen, J.-P.; Heikkilä, M.; Malinen, M.; Lee, H.-M.; Palvimo, J.J.; Wei, G.-H.; Myllyharju, J. A long hypoxia-inducible factor 3 isoform 2 is a transcription activator that regulates erythropoietin. Cell. Mol. Life Sci. 2019, 77, 3627–3642. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.A.; Simon, M.C. Biology of hypoxia-inducible factor-2alpha in development and disease. Cell Death Differ. 2008, 15, 628–634. [Google Scholar] [CrossRef]
- Zhao, C.X.; Luo, C.L.; Wu, X.H. Hypoxia promotes 786-O cells invasiveness and resistance to sorafenib via HIF-2alpha/COX-2. Med. Oncol. 2015, 32, 419. [Google Scholar] [CrossRef]
- Pawlus, M.R.; Wang, L.; Ware, K.; Hu, C.J. Upstream stimulatory factor 2 and hypoxia-inducible factor 2alpha (HIF2alpha) cooperatively activate HIF2 target genes during hypoxia. Mol. Cell. Biol. 2012, 32, 4595–4610. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Hill, H.; Christie, A.; Kim, M.S.; Holloman, E.; Pavia-Jimenez, A.; Homayoun, F.; Ma, Y.; Patel, N.; Yell, P.; et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 2016, 539, 112–117. [Google Scholar] [CrossRef] [Green Version]
- Metelo, A.M.; Noonan, H.R.; Li, X.; Jin, Y.; Baker, R.; Kamentsky, L.; Zhang, Y.; van Rooijen, E.; Shin, J.; Carpenter, A.E.; et al. Pharmacological HIF2alpha inhibition improves VHL disease-associated phenotypes in zebrafish model. J. Clin. Investig. 2015, 125, 1987–1997. [Google Scholar] [CrossRef] [Green Version]
- Lyberopoulou, A.; Venieris, E.; Mylonis, I.; Chachami, G.; Pappas, I.; Simos, G.; Bonanou, S.; Georgatsou, E. MgcRacGAP interacts with HIF-1alpha and regulates its transcriptional activity. Cell. Physiol. Biochem. 2007, 20, 995–1006. [Google Scholar] [CrossRef]
- Panda, A.C.; Martindale, J.L.; Gorospe, M. Polysome Fractionation to Analyze mRNA Distribution Profiles. Bio Protoc 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Befani, C.; Mylonis, I.; Gkotinakou, I.M.; Georgoulias, P.; Hu, C.J.; Simos, G.; Liakos, P. Cobalt stimulates HIF-1-dependent but inhibits HIF-2-dependent gene expression in liver cancer cells. Int. J. Biochem. Cell Biol. 2013, 45, 2359–2368. [Google Scholar] [CrossRef] [Green Version]
- Rankin, E.B.; Biju, M.P.; Liu, Q.; Unger, T.L.; Rha, J.; Johnson, R.S.; Simon, M.C.; Keith, B.; Haase, V.H. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J. Clin. Investig. 2007, 117, 1068–1077. [Google Scholar] [CrossRef]
- Hu, C.J.; Sataur, A.; Wang, L.; Chen, H.; Simon, M.C. The N-terminal transactivation domain confers target gene specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha. Mol. Biol. Cell 2007, 18, 4528–4542. [Google Scholar] [CrossRef] [Green Version]
- Karagiota, A.; Kourti, M.; Simos, G.; Mylonis, I. HIF-1α-derived cell-penetrating peptides inhibit ERK-dependent activation of HIF-1 and trigger apoptosis of cancer cells under hypoxia. Cell. Mol. Life Sci. 2018, 76, 809–825. [Google Scholar] [CrossRef]
- Mylonis, I.; Lakka, A.; Tsakalof, A.; Simos, G. The dietary flavonoid kaempferol effectively inhibits HIF-1 activity and hepatoma cancer cell viability under hypoxic conditions. Biochem. Biophys. Res. Commun. 2010, 398, 74–78. [Google Scholar] [CrossRef]
- Jones, G.; Prosser, D.E.; Kaufmann, M. Cytochrome P450-mediated metabolism of vitamin D. J. Lipid Res. 2014, 55, 13–31. [Google Scholar] [CrossRef] [Green Version]
- Angeli-Terzidou, A.-E.; Kechagia, E.; Pazaitou-Panayiotou, K.; Tsakalof, A. Vitamin D metabolizing enzymes: Expression and expression regulation by natural products in Huh7 cells. In Proceedings of the 69th Panhellenic Conference of the Hellenic Society for Biochemistry and Molecular Biology, Larissa, Greece, 23–25 November 2018; p. 143. [Google Scholar]
- Schito, M.; Hanna, D.; Zumla, A. Tuberculosis eradication versus control. Int. J. Infect. Dis 2017, 56, 10–13. [Google Scholar] [CrossRef] [Green Version]
- Semenza, G.L. Pharmacologic Targeting of Hypoxia-Inducible Factors. Annu. Rev. Pharm. Toxicol. 2019, 59, 379–403. [Google Scholar] [CrossRef]
- Wallace, E.M.; Rizzi, J.P.; Han, G.; Wehn, P.M.; Cao, Z.; Du, X.; Cheng, T.; Czerwinski, R.M.; Dixon, D.D.; Goggin, B.S.; et al. A Small-Molecule Antagonist of HIF2alpha Is Efficacious in Preclinical Models of Renal Cell Carcinoma. Cancer Res. 2016, 76, 5491–5500. [Google Scholar] [CrossRef] [Green Version]
- Wigerup, C.; Påhlman, S.; Bexell, D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol. Ther. 2016, 164, 152–169. [Google Scholar] [CrossRef] [Green Version]
- Giovannucci, E. The epidemiology of vitamin D and cancer incidence and mortality: A review (United States). Cancer Causes Control. 2005, 16, 83–95. [Google Scholar] [CrossRef]
- Giovannucci, E. Commentary: Vitamin D and colorectal cancer--twenty-five years later. Int. J. Epidemiol. 2006, 35, 222–224. [Google Scholar] [CrossRef]
- Ben-Shoshan, M.; Amir, S.; Dang, D.T.; Dang, L.H.; Weisman, Y.; Mabjeesh, N.J. 1alpha,25-dihydroxyvitamin D3 (Calcitriol) inhibits hypoxia-inducible factor-1/vascular endothelial growth factor pathway in human cancer cells. Mol. Cancer 2007, 6, 1433–1439. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.; Tang, B.; Sun, X. Development of Inhibitors Targeting Hypoxia-Inducible Factor 1 and 2 for Cancer Therapy. Yonsei Med. J. 2017, 58, 489–496. [Google Scholar] [CrossRef]
- Ge, X.; Wang, L.; Li, M.; Xu, N.; Yu, F.; Yang, F.; Li, R.; Zhang, F.; Zhao, B.; Du, J. Vitamin D/VDR signaling inhibits LPS-induced IFNγ and IL-1β in Oral epithelia by regulating hypoxia-inducible factor-1α signaling pathway. Cell Commun. Signal. 2019, 17. [Google Scholar] [CrossRef] [Green Version]
- Hubbi, M.E.; Hu, H.; Kshitiz; Ahmed, I.; Levchenko, A.; Semenza, G.L. Chaperone-mediated Autophagy Targets Hypoxia-inducible Factor-1α (HIF-1α) for Lysosomal Degradation. J. Biol. Chem. 2013, 288, 10703–10714. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Cao, J.; Zhao, L.; Zhu, S.; Chen, S.; Li, Y.; Zhao, B.; Zhao, T. PIM2 regulates stemness through phosphorylation of 4E-BP1. Sci. Bull. 2017, 62, 679–685. [Google Scholar] [CrossRef] [Green Version]
- Sang, N.; Stiehl, D.P.; Bohensky, J.; Leshchinsky, I.; Srinivas, V.; Caro, J. MAPK Signaling Up-regulates the Activity of Hypoxia-inducible Factors by Its Effects on p300. J. Biol. Chem. 2003, 278, 14013–14019. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Guo, X.; Liu, Z.; Zhao, X.; Feng, Y.; Song, S.; Cui, C.; Jiang, P. Vitamin D receptor activation influences the ERK pathway and protects against neurological deficits and neuronal death. Int. J. Mol. Med. 2017. [Google Scholar] [CrossRef]
- Zhao, J.-W.; Ping, J.-D.; Wang, Y.-F.; Liu, X.-N.; Li, N.; Hu, Z.-L.; Ming, L. Vitamin D suppress the production of vascular endothelial growth factor in mast cell by inhibiting PI3K/Akt/p38 MAPK/HIF-1α pathway in chronic spontaneous urticaria. Clin. Immunol. 2020, 215, 108444. [Google Scholar] [CrossRef]
- Huang, J.; Yang, G.; Huang, Y.; Zhang, S. Inhibitory effects of 1,25(OH)2D3 on the proliferation of hepatocellular carcinoma cells through the downregulation of HDAC2. Oncol. Rep. 2017, 38, 1845–1850. [Google Scholar] [CrossRef]
- Gingras, A.C.; Gygi, S.P.; Raught, B.; Polakiewicz, R.D.; Abraham, R.T.; Hoekstra, M.F.; Aebersold, R.; Sonenberg, N. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev. 1999, 13, 1422–1437. [Google Scholar] [CrossRef]
- Shveygert, M.; Kaiser, C.; Bradrick, S.S.; Gromeier, M. Regulation of eukaryotic initiation factor 4E (eIF4E) phosphorylation by mitogen-activated protein kinase occurs through modulation of Mnk1-eIF4G interaction. Mol. Cell. Biol. 2010, 30, 5160–5167. [Google Scholar] [CrossRef] [Green Version]
- Chiang, K.-C.; Chen, T.C. The Anti-cancer Actions of Vitamin, D. Anti-Cancer Agents Med. Chem. 2013, 13, 126–139. [Google Scholar] [CrossRef]
- Chung, I.; Han, G.; Seshadri, M.; Gillard, B.M.; Yu, W.d.; Foster, B.A.; Trump, D.L.; Johnson, C.S. Role of Vitamin D Receptor in the Antiproliferative Effects of Calcitriol in Tumor-Derived Endothelial Cells and Tumor Angiogenesis In vivo. Cancer Res. 2009, 69, 967–975. [Google Scholar] [CrossRef] [Green Version]
- Rius, J.; Guma, M.; Schachtrup, C.; Akassoglou, K.; Zinkernagel, A.S.; Nizet, V.; Johnson, R.S.; Haddad, G.G.; Karin, M. NF-κB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1α. Nature 2008, 453, 807–811. [Google Scholar] [CrossRef] [Green Version]
- van Uden, P.; Kenneth, N.S.; Rocha, S. Regulation of hypoxia-inducible factor-1α by NF-κB. Biochem. J. 2008, 412, 477–484. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhang, J.; Ge, X.; Du, J.; Deb, D.K.; Li, Y.C. Vitamin D receptor inhibits nuclear factor kappaB activation by interacting with IkappaB kinase beta protein. J. Biol. Chem. 2013, 288, 19450–19458. [Google Scholar] [CrossRef] [Green Version]
- Hoshida, Y.; Toffanin, S.; Lachenmayer, A.; Villanueva, A.; Minguez, B.; Llovet, J.M. Molecular classification and novel targets in hepatocellular carcinoma: Recent advancements. Semin. Liver Dis. 2010, 30, 35–51. [Google Scholar] [CrossRef] [Green Version]
- Min, L.; He, B.; Hui, L. Mitogen-activated protein kinases in hepatocellular carcinoma development. Semin. Cancer Biol. 2011, 21, 10–20. [Google Scholar] [CrossRef]
- Mylonis, I.; Simos, G. The Involvement of the ERK-Hypoxia-Angiogenesis Signaling Axis and HIF-1 in Hepatocellular Carcinoma. Hepatocell. Carcinoma Basic Res. 2012, 253–274. [Google Scholar] [CrossRef] [Green Version]
- Rosmorduc, O.; Housset, C. Hypoxia: A link between fibrogenesis, angiogenesis, and carcinogenesis in liver disease. Semin. Liver Dis. 2010, 30, 258–270. [Google Scholar] [CrossRef]
| 1 | F: CCCACTGAGGAGTCCAACATC |
| 2 | R: GGCCTTGGTGAGGTTTGATC |
| 3 | F: TTTTTCAAGCAGTAGGAATTGGA |
| 4 | F: TTCCTATTCACCAAGCTAAAGGAG |
| 5 | F: AGGCCGAGAATATCAGACG |
| 6 | F: CAGCTGACAGGAGGAGA |
| 7 | F: CTGTGGCTTCTGGCATACCT |
| 8 | F: TCTCAAGAAACAGCACGACACCC |
| 9 | F: CTCAACACGGGAAACCTCAC |
| 10 | R: CGCTCCACCAACTAAGAACG |











| Gene | Sequence (5′-3′) |
|---|---|
| VEGFA | F: CCCACTGAGGAGTCCAACATC R: GGCCTTGGTGAGGTTTGATC |
| HIF1A | F: TTTTTCAAGCAGTAGGAATTGGA R: GTGATGTAGTAGCTGCATGATCG |
| EPAS1 | F: TTCCTATTCACCAAGCTAAAGGAG R: GACTCCTCGAAGTTCTGATTCC |
| EPO | F: AGGCCGAGAATATCAGACG R: CCATCCTCTTCCAGGCATAGAAA |
| SERPINE1 | F: CAGCTGACAGGAGGAGA R: CCCATGAGCTCCTTGTACAGAT |
| PGK1 | F: CTGTGGCTTCTGGCATACCT R: CGAGTGACAGCCTCAGTATA |
| CYP24A1 | F: TCTCAAGAAACAGCACGACACCC R: GCACCGACTCAAAGGAACCCAAC |
| 18S | F: CTCAACACGGGAAACCTCAC R: CGCTCCACCAACTAAGAACG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gkotinakou, I.-M.; Kechagia, E.; Pazaitou-Panayiotou, K.; Mylonis, I.; Liakos, P.; Tsakalof, A. Calcitriol Suppresses HIF-1 and HIF-2 Transcriptional Activity by Reducing HIF-1/2α Protein Levels via a VDR-Independent Mechanism. Cells 2020, 9, 2440. https://doi.org/10.3390/cells9112440
Gkotinakou I-M, Kechagia E, Pazaitou-Panayiotou K, Mylonis I, Liakos P, Tsakalof A. Calcitriol Suppresses HIF-1 and HIF-2 Transcriptional Activity by Reducing HIF-1/2α Protein Levels via a VDR-Independent Mechanism. Cells. 2020; 9(11):2440. https://doi.org/10.3390/cells9112440
Chicago/Turabian StyleGkotinakou, Ioanna-Maria, Eleni Kechagia, Kalliopi Pazaitou-Panayiotou, Ilias Mylonis, Panagiotis Liakos, and Andreas Tsakalof. 2020. "Calcitriol Suppresses HIF-1 and HIF-2 Transcriptional Activity by Reducing HIF-1/2α Protein Levels via a VDR-Independent Mechanism" Cells 9, no. 11: 2440. https://doi.org/10.3390/cells9112440
APA StyleGkotinakou, I. -M., Kechagia, E., Pazaitou-Panayiotou, K., Mylonis, I., Liakos, P., & Tsakalof, A. (2020). Calcitriol Suppresses HIF-1 and HIF-2 Transcriptional Activity by Reducing HIF-1/2α Protein Levels via a VDR-Independent Mechanism. Cells, 9(11), 2440. https://doi.org/10.3390/cells9112440