Nuclear Morphological Remodeling in Human Granulocytes Is Linked to Prenylation Independently from Cytoskeleton
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. RNA Processing
2.3. Bioinformatics Analyses
2.4. Drug Treatments
2.5. Live-Cell Imaging
2.6. Image Analyses
2.7. Qualitative Evaluation of Nuclear Lobulation
2.8. Flow Cytometry Analyses
2.9. Molecular Modeling
2.10. Statistical Analyses
3. Results
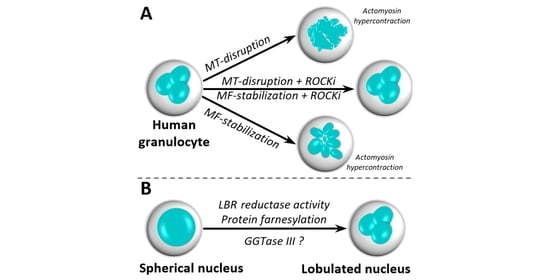
3.1. Lobular Structures in Segmented Nuclei Are Not Maintained by the Cytoskeleton
3.2. Nuclear Lobulation and Segmentation Progresses without the Involvement of Cytoskeleton
3.3. Transcriptomic Regulation of Sterol Metabolism Is Concurrent with Nuclear Remodeling
3.4. Intermediates of the Cholesterol Pathway and the Isoprenoid Branch Impact Nuclear Remodeling
3.5. Protein Prenylation Is Essential for Nuclear Remodeling
3.6. Putative Involvement of Novel GGTase III in Nuclear Remodeling
4. Discussion
4.1. The Role of Cytoskeleton in Nuclear Remodeling
4.2. The Role of Cholesterol Biosynthetic Pathway in Nuclear Remodeling
4.3. The Role of Prenyl-Transferases in Nuclear Remodeling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McGregor, A.L.; Hsia, C.R.; Lammerding, J. Squish and squeeze-the nucleus as a physical barrier during migration in confined environments. Curr. Opin. Cell Biol. 2016, 40, 32–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapisarda, V.; Malashchuk, I.; Asamaowei, I.E.; Poterlowicz, K.; Fessing, M.Y.; Sharov, A.A.; Karakesisoglou, I.; Botchkarev, V.A.; Mardaryev, A. p63 transcription factor regulates nuclear shape and expression of nuclear envelope-associated genes in epidermal keratinocytes. J. Investig. Dermatol. 2017, 137, 2157–2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bussolati, G.; Maletta, F.; Asioli, S.; Annaratone, L.; Sapino, A.; Marchiò, C. To be or not to be in a good shape: Diagnostic and clinical value of nuclear shape irregularities in thyroid and breast cancer. In Cancer Biology and the Nuclear Envelope: Recent Advances May Elucidate Past Paradoxes; Schirmer, E.C., de las Heras, J.I., Eds.; Springer: New York, NY, USA, 2014; pp. 101–121. [Google Scholar] [CrossRef]
- Fricker, M.; Hollinshead, M.; White, N.; Vaux, D. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J. Cell Biol. 1997, 136, 531–544. [Google Scholar] [CrossRef] [Green Version]
- Legartova, S.; Stixova, L.; Laur, O.; Kozubek, S.; Sehnalova, P.; Bartova, E. Nuclear structures surrounding internal lamin invaginations. J. Cell. Biochem. 2014, 115, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Bading, H. Nuclear calcium signalling in the regulation of brain function. Nat. Rev. Neurosci. 2013, 14, 593–608. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Hadipour-Lakmehsari, S.; Miyake, T.; Gramolini, A.O. Three-dimensional imaging reveals endo(sarco)plasmic reticulum-containing invaginations within the muscle nucleoplasm. Am. J. Physiol. Cell Physiol. 2017. [Google Scholar] [CrossRef]
- Drozdz, M.M.; Vaux, D.J. Shared mechanisms in physiological and pathological nucleoplasmic reticulum formation. Nucleus 2017, 8, 34–45. [Google Scholar] [CrossRef] [Green Version]
- Schoen, I.; Aires, L.; Ries, J.; Vogel, V. Nanoscale invaginations of the nuclear envelope: Shedding new light on wormholes with elusive function. Nucleus 2017, 8, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Campbell, M.S.; Lovell, M.A.; Gorbsky, G.J. Stability of nuclear segments in human neutrophils and evidence against a role for microfilaments or microtubules in their genesis during differentiation of HL60 myelocytes. J. Leukoc. Biol. 1995, 58, 659–666. [Google Scholar] [CrossRef]
- Neubert, E.; Meyer, D.; Rocca, F.; Gunay, G.; Kwaczala-Tessmann, A.; Grandke, J.; Senger-Sander, S.; Geisler, C.; Egner, A.; Schon, M.P.; et al. Chromatin swelling drives neutrophil extracellular trap release. Nat. Commun. 2018, 9, 3767. [Google Scholar] [CrossRef]
- Olins, A.L.; Olins, D.E. The mechanism of granulocyte nuclear shape determination: Possible involvement of the centrosome. Eur. J. Cell Biol. 2005, 84, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Jayo, A.; Malboubi, M.; Antoku, S.; Chang, W.; Ortiz-Zapater, E.; Groen, C.; Pfisterer, K.; Tootle, T.; Charras, G.; Gundersen, G.G.; et al. Fascin regulates nuclear movement and deformation in migrating cells. Dev. Cell 2016, 38, 371–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiam, H.R.; Vargas, P.; Carpi, N.; Crespo, C.L.; Raab, M.; Terriac, E.; King, M.C.; Jacobelli, J.; Alberts, A.S.; Stradal, T.; et al. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun. 2016, 7, 10997. [Google Scholar] [CrossRef] [PubMed]
- Wiggan, O.; Schroder, B.; Krapf, D.; Bamburg, J.R.; DeLuca, J.G. Cofilin regulates nuclear architecture through a Myosin-II dependent mechanotransduction module. Sci. Rep. 2017, 7, 40953. [Google Scholar] [CrossRef] [Green Version]
- Grespan, E.; Giobbe, G.G.; Badique, F.; Anselme, K.; Rühe, J.; Elvassore, N. Effect of geometrical constraints on human pluripotent stem cell nuclei in pluripotency and differentiation. Integr. Biol. Quant. Biosci. Nano Macro 2018, 10, 278–289. [Google Scholar] [CrossRef]
- Jorgens, D.M.; Inman, J.L.; Wojcik, M.; Robertson, C.; Palsdottir, H.; Tsai, W.T.; Huang, H.; Bruni-Cardoso, A.; Lopez, C.S.; Bissell, M.J.; et al. Deep nuclear invaginations are linked to cytoskeletal filaments—Integrated bioimaging of epithelial cells in 3D culture. J. Cell Sci. 2017, 130, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Gerlitz, G.; Reiner, O.; Bustin, M. Microtubule dynamics alter the interphase nucleus. Cell. Mol. Life Sci. CMLS 2013, 70, 1255–1268. [Google Scholar] [CrossRef]
- Paonessa, F.; Evans, L.D.; Solanki, R.; Larrieu, D.; Wray, S.; Hardy, J.; Jackson, S.P.; Livesey, F.J. Microtubules deform the nuclear membrane and disrupt nucleocytoplasmic transport in tau-mediated frontotemporal dementia. Cell Rep. 2019, 26, 582–593.e585. [Google Scholar] [CrossRef] [Green Version]
- Denais, C.M.; Gilbert, R.M.; Isermann, P.; McGregor, A.L.; te Lindert, M.; Weigelin, B.; Davidson, P.M.; Friedl, P.; Wolf, K.; Lammerding, J. Nuclear envelope rupture and repair during cancer cell migration. Science 2016, 352, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Raab, M.; Gentili, M.; de Belly, H.; Thiam, H.R.; Vargas, P.; Jimenez, A.J.; Lautenschlaeger, F.; Voituriez, R.; Lennon-Dumenil, A.M.; Manel, N.; et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 2016, 352, 359–362. [Google Scholar] [CrossRef]
- Audhya, A.; Desai, A.; Oegema, K. A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 2007, 178, 43–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciejowski, J.; Li, Y.; Bosco, N.; Campbell, P.J.; de Lange, T. Chromothripsis and kataegis induced by telomere crisis. Cell 2015, 163, 1641–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozler, J.; Nguyen, H.Q.; Rogers, G.C.; Bosco, G. Condensins exert force on chromatin-nuclear envelope tethers to mediate nucleoplasmic reticulum formation in Drosophila melanogaster. G3 (Bethesda) 2014, 5, 341–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olins, A.L.; Buendia, B.; Herrmann, H.; Lichter, P.; Olins, D.E. Retinoic acid induction of nuclear envelope-limited chromatin sheets in HL-60. Exp. Cell Res. 1998, 245, 91–104. [Google Scholar] [CrossRef]
- Olins, A.L.; Herrmann, H.; Lichter, P.; Kratzmeier, M.; Doenecke, D.; Olins, D.E. Nuclear envelope and chromatin compositional differences comparing undifferentiated and retinoic acid- and phorbol ester-treated HL-60 cells. Exp. Cell Res. 2001, 268, 115–127. [Google Scholar] [CrossRef]
- Zhu, Y.; Gong, K.; Denholtz, M.; Chandra, V.; Kamps, M.P.; Alber, F.; Murre, C. Comprehensive characterization of neutrophil genome topology. Genes Dev. 2017, 31, 141–153. [Google Scholar] [CrossRef] [Green Version]
- Karni, R.J.; Wangh, L.J.; Sanchez, A.J. Nonrandom location and orientation of the inactive X chromosome in human neutrophil nuclei. Chromosoma 2001, 110, 267–274. [Google Scholar] [CrossRef]
- Sanchez, J.; Karni, R.; Wangh, L. Fluorescent in situ Hybridization (FISH) Analysis of the Relationship between Chromosome Location and Nuclear Morphology in Human Neutrophils. Chromosoma 1997, 106, 168–177. [Google Scholar] [CrossRef]
- Gravemann, S.; Schnipper, N.; Meyer, H.; Vaya, A.; Nowaczyk, M.J.; Rajab, A.; Hofmann, W.K.; Salewsky, B.; Tonnies, H.; Neitzel, H.; et al. Dosage effect of zero to three functional LBR-genes in vivo and in vitro. Nucleus 2010, 1, 179–189. [Google Scholar] [CrossRef]
- Rowat, A.C.; Jaalouk, D.E.; Zwerger, M.; Ung, W.L.; Eydelnant, I.A.; Olins, D.E.; Olins, A.L.; Herrmann, H.; Weitz, D.A.; Lammerding, J. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J. Biol. Chem. 2013, 288, 8610–8618. [Google Scholar] [CrossRef] [Green Version]
- Schnipper, N.; Stassen, H.H.; Kallinich, T.; Sperling, K.; Hoffmann, K. Image analysis of neutrophil nuclear morphology: Learning about phenotypic range and its reliable analysis from patients with pelger-Huet-anomaly and treated with colchicine. Cytom. Part B Clin. Cytom. 2017, 92, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Martewicz, S.; Luni, C.; Serena, E.; Pavan, P.; Chen, H.V.; Rampazzo, A.; Elvassore, N. Transcriptomic characterization of a human in vitro model of arrhythmogenic cardiomyopathy under topological and mechanical stimuli. Ann. Biomed. Eng. 2019, 47, 852–865. [Google Scholar] [CrossRef] [PubMed]
- Pfisterer, K.; Jayo, A.; Parsons, M. Control of nuclear organization by F-actin binding proteins. Nucleus 2017, 8, 126–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatau, S.B.; Hale, C.M.; Stewart-Hutchinson, P.J.; Patel, M.S.; Stewart, C.L.; Searson, P.C.; Hodzic, D.; Wirtz, D. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA 2009, 106, 19017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maninova, M.; Caslavsky, J.; Vomastek, T. The assembly and function of perinuclear actin cap in migrating cells. Protoplasma 2017. [Google Scholar] [CrossRef] [PubMed]
- Isogai, T.; Innocenti, M. New nuclear and perinuclear functions of formins. Biochem. Soc. Trans. 2016, 44, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Alisafaei, F.; Jokhun, D.S.; Shivashankar, G.V.; Shenoy, V.B. Regulation of nuclear architecture, mechanics, and nucleocytoplasmic shuttling of epigenetic factors by cell geometric constraints. Proc. Natl. Acad. Sci. USA 2019, 116, 13200–13209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, F.-Y.; Haley, S.C.; Zidovska, A. On the origin of shape fluctuations of the cell nucleus. Proc. Natl. Acad. Sci. USA 2017, 114, 10338. [Google Scholar] [CrossRef] [Green Version]
- Makhija, E.; Jokhun, D.S.; Shivashankar, G.V. Nuclear deformability and telomere dynamics are regulated by cell geometric constraints. Proc. Natl. Acad. Sci. USA 2016, 113, E32. [Google Scholar] [CrossRef] [Green Version]
- Ramdas, N.M.; Shivashankar, G.V. Cytoskeletal control of nuclear morphology and chromatin organization. J. Mol. Biol. 2015, 427, 695–706. [Google Scholar] [CrossRef]
- Chang, Y.C.; Nalbant, P.; Birkenfeld, J.; Chang, Z.F.; Bokoch, G.M. GEF-H1 couples nocodazole-induced microtubule disassembly to cell contractility via RhoA. Mol. Biol. Cell 2008, 19, 2147–2153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolodney, M.S.; Elson, E.L. Contraction due to microtubule disruption is associated with increased phosphorylation of myosin regulatory light chain. Proc. Natl. Acad. Sci. USA 1995, 92, 10252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krendel, M.; Zenke, F.T.; Bokoch, G.M. Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat. Cell Biol. 2002, 4, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Patteson, A.E.; Pogoda, K.; Byfield, F.J.; Mandal, K.; Ostrowska-Podhorodecka, Z.; Charrier, E.E.; Galie, P.A.; Deptuła, P.; Bucki, R.; McCulloch, C.A.; et al. Loss of vimentin enhances cell motility through small confining spaces. Small 2019, 15, 1903180. [Google Scholar] [CrossRef]
- Patteson, A.E.; Vahabikashi, A.; Pogoda, K.; Adam, S.A.; Mandal, K.; Kittisopikul, M.; Sivagurunathan, S.; Goldman, A.; Goldman, R.D.; Janmey, P.A. Vimentin protects cells against nuclear rupture and DNA damage during migration. J. Cell Biol. 2019, 218, 4079–4092. [Google Scholar] [CrossRef]
- Biskou, O.; Casanova, V.; Hooper, K.M.; Kemp, S.; Wright, G.P.; Satsangi, J.; Barlow, P.G.; Stevens, C. The type III intermediate filament vimentin regulates organelle distribution and modulates autophagy. PLoS ONE 2019, 14, e0209665. [Google Scholar] [CrossRef]
- Olins, A.L.; Herrmann, H.; Lichter, P.; Olins, D.E. Retinoic acid differentiation of HL-60 cells promotes cytoskeletal polarization. Exp. Cell Res. 2000, 254, 130–142. [Google Scholar] [CrossRef]
- Olins, A.L.; Olins, D.E. Cytoskeletal influences on nuclear shape in granulocytic HL-60 cells. BMC Cell Biol. 2004, 5, 30. [Google Scholar] [CrossRef] [Green Version]
- Shahoei, S.H.; Nelson, E.R. Nuclear receptors, cholesterol homeostasis and the immune system. J. Steroid Biochem. Mol. Biol. 2019, 191, 105364. [Google Scholar] [CrossRef]
- Yachnin, S.; Larson, R.A.; West, E.J. Rates of cholesterol biosynthesis are related to early differentiation in acute non-lymphocytic leukaemia cells. Br. J. Haematol. 1983, 54, 459–466. [Google Scholar] [CrossRef]
- Yachnin, S.; Toub, D.B.; Mannickarottu, V. Divergence in cholesterol biosynthetic rates and 3-hydroxy-3-methylglutaryl-CoA reductase activity as a consequence of granulocyte versus monocyte-macrophage differentiation in HL-60 cells. Proc. Natl. Acad. Sci. USA 1984, 81, 894–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, K.; Dreger, C.K.; Olins, A.L.; Olins, D.E.; Shultz, L.D.; Lucke, B.; Karl, H.; Kaps, R.; Muller, D.; Vaya, A.; et al. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huet anomaly). Nat. Genet. 2002, 31, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Lagace, T.A.; Ridgway, N.D. The rate-limiting enzyme in phosphatidylcholine synthesis regulates proliferation of the nucleoplasmic reticulum. Mol. Biol. Cell 2005, 16, 1120–1130. [Google Scholar] [CrossRef] [PubMed]
- Drozdz, M.M.; Jiang, H.; Pytowski, L.; Grovenor, C.; Vaux, D.J. Formation of a nucleoplasmic reticulum requires de novo assembly of nascent phospholipids and shows preferential incorporation of nascent lamins. Sci. Rep. 2017, 7, 7454. [Google Scholar] [CrossRef] [Green Version]
- Goulbourne, C.N.; Malhas, A.N.; Vaux, D.J. The induction of a nucleoplasmic reticulum by prelamin A accumulation requires CTP: Phosphocholine cytidylyltransferase-α. J. Cell Sci. 2011, 124, 4253–4266. [Google Scholar] [CrossRef] [Green Version]
- Martinovic, T.; Ciric, D.; Pantic, I.; Lalic, K.; Rasulic, I.; Despotovic, S.; Lalic, I.; Djuricic, D.; Bumbasirevic, V.; Kravic-Stevovic, T. Unusual shape and structure of lymphocyte nuclei is linked to hyperglycemia in type 2 diabetes patients. Tissue Cell 2018, 52, 92–100. [Google Scholar] [CrossRef]
- Gatticchi, L.; Cerra, B.; Scarpelli, P.; Macchioni, L.; Sebastiani, B.; Gioiello, A.; Roberti, R. Selected cholesterol biosynthesis inhibitors produce accumulation of the intermediate FF-MAS that targets nucleus and activates LXRα in HepG2 cells. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 842–852. [Google Scholar] [CrossRef]
- Lindenthal, B.; Holleran, A.L.; Aldaghlas, T.A.; Ruan, B.; Schroepfer, G.J.; Wilson, W.K.; Kelleher, J.K. Progestins block cholesterol synthesis to produce meiosis-activating sterols. FASEB J. 2001, 15, 775–784. [Google Scholar] [CrossRef]
- Saito, M.; Nojiri, H.; Ogino, H.; Yuo, A.; Ogura, H.; Itoh, M.; Tomita, K.; Ogawa, T.; Nagai, Y.; Kitagawa, S. Synthetic sialyl glycolipids (sialo-cholesterol and sialo-diglyceride) induce granulocytic differentiation of human myelogenous leukemia cell line HL-60. FEBS Lett. 1990, 271, 85–88. [Google Scholar] [CrossRef] [Green Version]
- Gregorio-King, C.C.; Collier, F.M.; Bolton, K.A.; Ferguson, M.; Hosking, J.B.; Collier, G.R.; Kirkland, M.A. Effect of oxysterols on hematopoietic progenitor cells. Exp. Hematol. 2002, 30, 670–678. [Google Scholar] [CrossRef]
- Pytowski, L.; Drozdz, M.M.; Jiang, H.; Hernandez, Z.; Kumar, K.; Knott, E.; Vaux, D.J. Nucleoplasmic reticulum formation in human endometrial cells is steroid hormone responsive and recruits nascent components. Int. J. Mol. Sci. 2019, 20, 5839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.C.; Liu, T.Y.; Huang, S.P.; Ho, C.T.; Huang, T.C. Differentiation and apoptosis induction by lovastatin and gamma-tocotrienol in HL-60 cells via Ras/ERK/NF-kappaB and Ras/Akt/NF-kappaB signaling dependent down-regulation of glyoxalase 1 and HMG-CoA reductase. Cell. Signal. 2015, 27, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Dimitroulakos, J.; Thai, S.; Wasfy, G.H.; Hedley, D.W.; Minden, M.D.; Penn, L.Z. Lovastatin induces a pronounced differentiation response in acute myeloid leukemias. Leuk. Lymphoma 2000, 40, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.; Clutterbuck, R.D.; Powles, R.L.; Catovsky, D.; Millar, J.L. A comparison of the effect of the 3-Hydroxy-3-Methylglutaryl Coenzyme A (HMG-CoA) reductase inhibitors simvastatin, lovastatin and pravastatin on leukaemic and normal bone marrow progenitors. Leuk. Lymphoma 1997, 24, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gardner, J.P.; Kheir, A.; Uskokovic, M.R.; Studzinski, G.P. Synergistic induction of HL60 cell differentiation by ketoconazole and 1-desoxy analogues of vitamin D3. J. Natl. Cancer Inst. 1997, 89, 1199–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, C.; Martin, M.; Gomez-Coronado, D.; Lasuncion, M.A. Effects of distal cholesterol biosynthesis inhibitors on cell proliferation and cell cycle progression. J. Lipid Res. 2005, 46, 920–929. [Google Scholar] [CrossRef] [Green Version]
- Nikolakaki, E.; Mylonis, I.; Giannakouros, T. Lamin B receptor: Interplay between structure, function and localization. Cells 2017, 6, 28. [Google Scholar] [CrossRef]
- Bennati, A.M.; Castelli, M.; Della Fazia, M.A.; Beccari, T.; Caruso, D.; Servillo, G.; Roberti, R. Sterol dependent regulation of human TM7SF2 gene expression: Role of the encoded 3beta-hydroxysterol Delta14-reductase in human cholesterol biosynthesis. Biochim. Biophys. Acta 2006, 1761, 677–685. [Google Scholar] [CrossRef]
- Tsai, P.-L.; Zhao, C.; Turner, E.; Schlieker, C. The lamin B receptor is essential for cholesterol synthesis and perturbed by disease-causing mutations. eLife 2016, 5, e16011. [Google Scholar] [CrossRef]
- Bennati, A.M.; Schiavoni, G.; Franken, S.; Piobbico, D.; Della Fazia, M.A.; Caruso, D.; De Fabiani, E.; Benedetti, L.; Cusella De Angelis, M.G.; Gieselmann, V.; et al. Disruption of the gene encoding 3beta-hydroxysterol Delta-reductase (Tm7sf2) in mice does not impair cholesterol biosynthesis. FEBS J. 2008, 275, 5034–5047. [Google Scholar] [CrossRef]
- Subramanian, G.; Chaudhury, P.; Malu, K.; Fowler, S.; Manmode, R.; Gotur, D.; Zwerger, M.; Ryan, D.; Roberti, R.; Gaines, P. Lamin B receptor regulates the growth and maturation of myeloid progenitors via its sterol reductase domain: Implications for cholesterol biosynthesis in regulating myelopoiesis. J. Immunol. 2012, 188, 85–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moutinho, M.; Nunes, M.J.; Rodrigues, E. The mevalonate pathway in neurons: It’s not just about cholesterol. Exp. Cell Res. 2017, 360, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, J.; Kimura, S.; Segawa, H.; Kobayashi, Y.; Yoshikawa, T.; Urasaki, Y.; Ueda, T.; Enjo, F.; Tokuda, H.; Ottmann, O.G.; et al. The third-generation bisphosphonate zoledronate synergistically augments the anti-Ph+ leukemia activity of imatinib mesylate. Blood 2003, 102, 2229–2235. [Google Scholar] [CrossRef] [PubMed]
- Prufert, K.; Alsheimer, M.; Benavente, R.; Krohne, G. The myristoylation site of meiotic lamin C2 promotes local nuclear membrane growth and the formation of intranuclear membranes in somatic cultured cells. Eur. J. Cell Biol. 2005, 84, 637–646. [Google Scholar] [CrossRef]
- Prufert, K.; Vogel, A.; Krohne, G. The lamin CxxM motif promotes nuclear membrane growth. J. Cell Sci. 2004, 117, 6105–6116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrovsky, R.; Krohne, G.; Großhans, J. Overexpression of the lamina proteins Lamin and Kugelkern induces specific ultrastructural alterations in the morphology of the nuclear envelope of intestinal stem cells and enterocytes. Eur. J. Cell Biol. 2018, 97, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Polychronidou, M.; Hellwig, A.; Grosshans, J. Farnesylated nuclear proteins Kugelkern and lamin Dm0 affect nuclear morphology by directly interacting with the nuclear membrane. Mol. Biol. Cell 2010, 21, 3409–3420. [Google Scholar] [CrossRef] [Green Version]
- Verstraeten, V.L.; Peckham, L.A.; Olive, M.; Capell, B.C.; Collins, F.S.; Nabel, E.G.; Young, S.G.; Fong, L.G.; Lammerding, J. Protein farnesylation inhibitors cause donut-shaped cell nuclei attributable to a centrosome separation defect. Proc. Natl. Acad. Sci. USA 2011, 108, 4997–5002. [Google Scholar] [CrossRef] [Green Version]
- Trigueros-Motos, L.; Gonzalez, J.M.; Rivera, J.; Andres, V. Hutchinson-Gilford progeria syndrome, cardiovascular disease and oxidative stress. Front. Biosci. Sch. Ed. 2011, 3, 1285–1297. [Google Scholar] [CrossRef] [Green Version]
- Bartova, E.; Kozubek, S.; Jirsova, P.; Kozubek, M.; Lukasova, E.; Skalnikova, M.; Cafourkova, A.; Koutna, I.; Pasekova, R. Higher-order chromatin structure of human granulocytes. Chromosoma 2001, 110, 360–370. [Google Scholar] [CrossRef]
- Ellenberg, J.; Siggia, E.D.; Moreira, J.E.; Smith, C.L.; Presley, J.F.; Worman, H.J.; Lippincott-Schwartz, J. Nuclear membrane dynamics and reassembly in living cells: Targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 1997, 138, 1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volkova, E.G.; Kurchashova, S.Y.; Polyakov, V.Y.; Sheval, E.V. Self-organization of cellular structures induced by the overexpression of nuclear envelope proteins: A correlative light and electron microscopy study. Microscopy 2010, 60, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Bokoch, G.M.; Prossnitz, V. Isoprenoid metabolism is required for stimulation of the respiratory burst oxidase of HL-60 cells. J. Clin. Investig. 1992, 89, 402–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiemer, A.J.; Wiemer, D.F.; Hohl, R.J. Geranylgeranyl diphosphate synthase: An emerging therapeutic target. Clin. Pharmacol. Ther. 2011, 90, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Lobell, R.B.; Liu, D.; Buser, C.A.; Davide, J.P.; DePuy, E.; Hamilton, K.; Koblan, K.S.; Lee, Y.; Mosser, S.; Motzel, S.L.; et al. Preclinical and clinical pharmacodynamic assessment of L-778,123, a dual inhibitor of farnesyl:protein transferase and geranylgeranyl:protein transferase type-I. Mol. Cancer Ther. 2002, 1, 747–758. [Google Scholar] [PubMed]
- Reid, T.S.; Long, S.B.; Beese, L.S. Crystallographic analysis reveals that anticancer clinical candidate L-778,123 inhibits protein farnesyltransferase and geranylgeranyltransferase-I by different binding modes. Biochemistry 2004, 43, 9000–9008. [Google Scholar] [CrossRef]
- Shirakawa, R.; Goto-Ito, S.; Goto, K.; Wakayama, S.; Kubo, H.; Sakata, N.; Trinh, D.A.; Yamagata, A.; Sato, Y.; Masumoto, H.; et al. A SNARE geranylgeranyltransferase essential for the organization of the Golgi apparatus. EMBO J. 2020, 39, e104120. [Google Scholar] [CrossRef]
- Kuchay, S.; Wang, H.; Marzio, A.; Jain, K.; Homer, H.; Fehrenbacher, N.; Philips, M.R.; Zheng, N.; Pagano, M. GGTase3 is a newly identified geranylgeranyltransferase targeting a ubiquitin ligase. Nat. Struct. Mol. Biol. 2019, 26, 628–636. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martewicz, S.; Luni, C.; Zhu, X.; Cui, M.; Hu, M.; Qu, S.; Buratto, D.; Yang, G.; Grespan, E.; Elvassore, N. Nuclear Morphological Remodeling in Human Granulocytes Is Linked to Prenylation Independently from Cytoskeleton. Cells 2020, 9, 2509. https://doi.org/10.3390/cells9112509
Martewicz S, Luni C, Zhu X, Cui M, Hu M, Qu S, Buratto D, Yang G, Grespan E, Elvassore N. Nuclear Morphological Remodeling in Human Granulocytes Is Linked to Prenylation Independently from Cytoskeleton. Cells. 2020; 9(11):2509. https://doi.org/10.3390/cells9112509
Chicago/Turabian StyleMartewicz, Sebastian, Camilla Luni, Xi Zhu, Meihua Cui, Manli Hu, Siqi Qu, Damiano Buratto, Guang Yang, Eleonora Grespan, and Nicola Elvassore. 2020. "Nuclear Morphological Remodeling in Human Granulocytes Is Linked to Prenylation Independently from Cytoskeleton" Cells 9, no. 11: 2509. https://doi.org/10.3390/cells9112509
APA StyleMartewicz, S., Luni, C., Zhu, X., Cui, M., Hu, M., Qu, S., Buratto, D., Yang, G., Grespan, E., & Elvassore, N. (2020). Nuclear Morphological Remodeling in Human Granulocytes Is Linked to Prenylation Independently from Cytoskeleton. Cells, 9(11), 2509. https://doi.org/10.3390/cells9112509