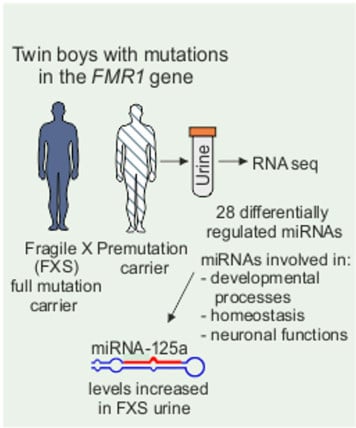
Urine microRNA Profiling Displays miR-125a Dysregulation in Children with Fragile X Syndrome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Urine Samples
2.2. RNA Extraction and Quantitative Real-Time PCR
2.3. Deep Sequencing
2.4. Analysis of Data
3. Results
3.1. Profiling of FXS Urine miRNAs by Deep Sequencing
3.2. Increased miR-125a in Urine of Children with FXS
3.3. Age-Dependent Dysregulation of miR-125a in Urine of Children with FXS
3.4. Target Gene and Pathway Analysis of miRNAs Regulated Differentially in FXS Urine
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartel, D. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.; Feinbaum, R.; Ambros, V. The c. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Cheng, L.; Sun, X.; Scicluna, B.; Coleman, B.; Hill, A. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014, 86, 433–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, J.; Baxter, D.; Zhang, S.; Huang, D.; Huang, K.; Lee, M.; Galas, D.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Kenny, P. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012, 35, 325–334. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, S. An unconventional role for miRNAs: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012, 15, 827–835. [Google Scholar] [CrossRef]
- Xihua, L. Circulating miR-143-3p inhibition protects against insulin resistance in Metabolic Syndrome via targeting of the insulin-like growth factor 2 receptor. Transl. Res. 2018, 205, 33–43. [Google Scholar] [CrossRef] [Green Version]
- Hagerman, R.; Berry-Kravis, E.; Kaufmann, W.; Ono, M.; Tartaglia, N.; Lachiewicz, A.; Kronk, R.; Delahunty, C.; Hessl, D.; Visootsak, J.; et al. Advances in the treatment of fragile X syndrome. Pediatrics 2009, 123, 378–390. [Google Scholar] [CrossRef] [Green Version]
- Terraciano, A.; Chiurazza, P.; Neri, G. Fragile X syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2005, 137, 32–37. [Google Scholar] [CrossRef]
- Berry-Kravis, E. Epilepsy in fragile X syndrome. Dev. Med. Child. Neurol. 2002, 44, 724–728. [Google Scholar] [CrossRef]
- Pieretti, M.; Zhang, F.; Fu, Y.-H.; Warren, S.T.; Oostra, B.A.; Caskey, C.T.; Nelson, D.L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 1991, 66, 817–822. [Google Scholar] [CrossRef]
- Hagerman, R.J.; Hagerman, P.J. The fragile X premutation: Into the phenotypic fold. Curr. Opin Genet. Dev. 2002, 12, 278–283. [Google Scholar] [CrossRef]
- Hagerman, P.; Hagerman, R. Fragile X-associated tremor/ataxia syndrome (FXTAS). Mental Ret. Dev. Disabil. Res. Rev. 2004, 10, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Khandjian, E.W.; Fortin, A.; Thibodeau, A.; Tremblay, S.; Cote, F.; Devys, D.; Mandel, J.L.; Rousseau, F. A heterogeneous set of FMR1 proteins is widely distributed in mouse tissues and is modulated in cell culture. Hum. Mol. Genet. 1995, 4, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Slegtenhorst-Eegdeman, K.; de Rooij, D.; Verhoef-Post, M.; van de Kant, H.; Bakker, C.; Oostra, B.; Grootegoed, J.; Themmen, A. Macroorchidism in fmr1 knockout mice is caused by increased Sertoli cell proliferation during testicular development. Endocrinology 1998, 39, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Edbauer, D.; Neilson, J.; Foster, K.; Wang, C.; Seeburg, D.; Batterton, M.; Tada, T.; Dolan, B.; Sharp, P.; Sheng, M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 2010, 65, 373–384. [Google Scholar] [CrossRef] [Green Version]
- Gong, X.; Wang, Y.; Zeng, J.; Li, S.; Luo, Y. Computational identification and experimental validation of microRNAs binding to the fragile X syndrome gene Fmr1. Neurochem. Res. 2015, 40, 109–117. [Google Scholar] [CrossRef]
- DeMarco, B.; Stefanovic, S.; Williams, A.; Moss, K.; Anderson, B.; Bassell, G.; Mihailescu, M. FMRP-G-quadruplex mRNA-miR-125a interactions: Implications for miR-125a mediated translation regulation of PSD-95 mRNA. PLoS ONE 2019, 14, e0217275. [Google Scholar] [CrossRef]
- Muddashetty, R.; Nalavadi, V.; Gross, C.; Yao, X.; Xing, L.; Laur, O.; Warren, S.; Bassell, G. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell 2011, 42, 673–688. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Wu, N.; Wang, L.; Li, J. Circulating microRNAs: A novel class of potential biomarkers for diagnosing and prognosing central nervous system diseases. Cell Mol. Neurobiol. 2013, 33, 601–613. [Google Scholar] [CrossRef]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qureshi, R.; Sacan, A. A novel method for the normalization of microRNA RT-PCR data. BMC Med. Genomics 2013, 6 (Suppl. 1), 14. [Google Scholar] [CrossRef] [Green Version]
- Love, M.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for rna-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.-P.; Lee, C.-Y.; Tsai, M.-H.; Chiu, Y.-C.; Hsiao, C.; Lai, L.-C.; Chuang, E. miRSystem: An integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS ONE 2012, 7, e42390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Sherman, B.; Lempicki, R. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, 457–462. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Lin, K.; Chen, Y. Diverse functions of miRNA-125a family in different cell contexts. Hematol. Oncol. 2013, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- Reijerkerk, A.; Lopez-Ramirez, M.; van Het Hof, B.; Drexhage, J.; Kamphuis, W.; Kooij, G.; Vos, J.; van der Pouw Kraan, T.; van Zonneveld, A.; Horrevoets, A.; et al. MicroRNAs regulate human brain endothelial cell-barrier function in inflammation: Implications for multiple sclerosis. J. Neurosci 2013, 33, 6857–6863. [Google Scholar] [CrossRef] [Green Version]
- Dakir, E.-H.; Mollinedo, F. Genome-wide miRNA profiling and pivotal roles of miRs 125a-5p and 17-92 cluster in human neutrophil maturation and differentiation of acute myeloid leukemia cells. Oncotarget 2019, 10, 5313–5331. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Yang, P.; Xiong, Q.; Song, X.; Yang, X.; Liu, L.; Yuan, W.; Rui, Y. MicroRNA-125a/b-5p inhibits endothelin-1 expression in vascular endothelial cells. J. Hypertens. 2010, 8, 1646–1654. [Google Scholar] [CrossRef]
- Niu, W.; Sun, B.; Li, M.; Cui, J.; Huang, J.; Zhang, L. TLR-4/microRNA-125a/NF-κB signaling modulates the immune response to Mycobacterium tuberculosis infection. Cell Cycle 2018, 17, 1931–1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Huang, X.; Lu, S.; Deng, H.; Gan, H.; Huang, R.; Zhang, B. miRNA-125a modulates autophagy of thyroiditis through PI3K/Akt/mTOR signaling pathway. Exp. Ther. Med. 2019, 17, 2465–2472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, H.; Zhu, W.; Mei, L.; Lu, Z. Prognostic and clinicopathologic significance of MicroRNA-125a-5p in cancers: A meta-analysis. Medicine 2019, 98, e16685. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, L.; Yan, G.; Zhang, W.; Huan, Z.; Li, J. miR-125a-5p alleviates dysfunction and inflammation of pentylenetetrazol-induced epilepsy through targeting calmodulin-dependent protein kinase IV (CAMK4). Curr. Neurovasc. Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.; Remaley, A. Lipid-based carriers of microRNAs and intercellular communication. Curr. Opin. Lipidol. 2012, 23, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Bray, S.M.; Warren, S.T. New perspectives on the biology of fragile X syndrome. Curr. Opin. Genet. Dev. 2012, 22, 256–263. [Google Scholar] [CrossRef] [Green Version]
- Zen, K.; Zhang, C. Circulating microRNAs: A novel class of biomarkers to diagnose and monitor human cancers. Med. Res. Rev. 2012, 32, 326–348. [Google Scholar] [CrossRef]
- Liang, H.; Gong, F.; Zhang, S.; Zhang, C.; Zen, K.; Chen, X. The origin, function, and diagnostic potential of extracellular microRNAs in human body fluids. Wiley Interdiscip. Rev. 2013, 5, 285–300. [Google Scholar] [CrossRef]
- Chen, X.; Liang, H.; Zhang, J.; Zen, K.; Zhang, C. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012, 22, 125–132. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjöstrand, M.; Lee, J.; Lötvall, J. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Arroyo, J.; Chevillet, J.; Kroh, E.; Ruf, I.; Pritchard, C.; Gibson, D.; Mitchell, P.; Bennett, C.; Pogosova-Agadjanyan, E.; Stirewalt, D.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez-Mora, M.; Rodriguez-Revenga, L.; Madrigal, I.; Torres-Silva, F.; Mateu-Huertas, E.; Lizano, E.; Friedländer, M.; Martí, E.; Estivill, X.; Milà, M. MicroRNA expression profiling in blood from fragile X-associated tremor/ataxia syndrome patients. Genes Brain Behav. 2013, 12, 595–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mundalil Vasu, M.; Anitha, A.; Thanseem, I.; Suzuki, K.; Yamada, K.; Takahashi, T.; Wakuda, T.; Iwata, K.; Tsujii, M.; Sugiyama, T.; et al. Serum microRNA profiles in children with autism. Mol. Autism 2014, 5, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boissart, C.; Nissan, X.; Giraud-Triboult, K.; Peschanski, M.; Benchoua, A. miR-125 potentiates early neural specification of human embryonic stem cells. Development 2012, 139, 1247–1257. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.-L. microRNAs and Fragile X Syndrome. Adv. Exp. Med. Biol. 2015, 888, 107–121. [Google Scholar] [CrossRef]
- Ziats, M.; Rennert, O. Identification of differentially expressed microRNAs across the developing human brain. Mol. Psychiatry 2014, 19, 848–852. [Google Scholar] [CrossRef] [Green Version]
- Schumann, C.; Sharp, F.; Ander, B.; Stamova, B. Possible sexually dimorphic role of miRNA and other sncRNA in ASD brain. Mol. Autism 2017, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Abu-Elneel, K.; Liu, T.; Gazzaniga, F.; Nishimura, Y.; Wall, D.; Geschwind, D.; Lao, K.; Kosik, K. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics 2008, 9, 153–161. [Google Scholar] [CrossRef]
- Gurwitz, D. Genomics and the future of psychopharmacology: MicroRNAs offer novel therapeutics. Dialogues Clin. Neurosci. 2019, 21, 131–148. [Google Scholar]
- Camkurt, M.; Karababa, İ.; Erdal, M.; Kandemir, S.; Fries, G.; Bayazıt, H.; Ay, M.; Kandemir, H.; Ay, Ö.; Coşkun, S.; et al. MicroRNA dysregulation in manic and euthymic patients with bipolar disorder. J. Affect. Disord 2019, 12, 284–290. [Google Scholar] [CrossRef]




| Subject/phenotype | Age; Years | Gender | Repeat Length | Medication |
|---|---|---|---|---|
| FXS1 | 4 | male | mosaic | |
| FXS2 | 5 | male | >200 | risperidone, methyphenidate |
| FXS3 | 6 | male | 86/520 | methylphenidate |
| FXS4 | 6 | male | >200 | |
| FXS5 | 8 | male | >200 | |
| FXS6 | 8 | male | 300 | oxcarbatzepine |
| FXS7 | 13 | male | >200 | methylphenidate |
| FXS8 | 14 | female | 34/500 | |
| FXS9 | 17 | male | 300 |
| Target Gene | Gene Description | Observed Number of miRNA |
|---|---|---|
| STARD13 | StAR-related lipid transfer (START) domain containing 13 | 5 |
| GABRB2 | gamma-aminobutyric acid (GABA) A receptor, beta 2 | 5 |
| BDNF | brain-derived neurotrophic factor | 4 |
| CECR6 | cat eye syndrome chromosome region, candidate 6 | 4 |
| CNOT6 | CCR4-NOT transcription complex, subunit 6 | 4 |
| CRK | v-crk avian sarcoma virus CT10 oncogene homolog | 4 |
| DPF2 | D4, zinc and double PHD fingers family 2 | 4 |
| DVL3 | dishevelled segment polarity protein 3 | 4 |
| ELOVL6 | ELOVL fatty acid elongase 6 | 4 |
| EPHA4 | EPH receptor A4 | 4 |
| FIGN | fidgetin | 4 |
| BCL2L2 | BCL2-like 2 | 4 |
| FOXP1 | forkhead box P1 | 4 |
| GRIN3A | glutamate receptor, ionotropic, N-methyl-D-aspartate 3A | 4 |
| IGF2BP2 | insulin-like growth factor 2 mRNA binding protein 2 | 4 |
| IL1RAPL1 | interleukin 1 receptor accessory protein-like 1 | 4 |
| KCNA1 | potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) | 4 |
| MTF1 | metal-regulatory transcription factor 1 | 4 |
| MYCBP | MYC binding protein | 4 |
| ONECUT2 | one cut homeobox 2 | 4 |
| RASL10B | RAS-like, family 10, member B | 4 |
| SORT1 | sortilin 1 | 4 |
| Database | Pathway | Target genes | Score |
|---|---|---|---|
| REACTOME | Developmental Biology | 105 | 3.663 |
| REACTOME | Axon guidance | 72 | 3.588 |
| KEGG | MAPK signaling pathway | 67 | 3.212 |
| REACTOME | L1CAM interactions | 32 | 3.050 |
| KEGG | Pathways in cancer | 73 | 2.827 |
| BIOCARTA | Biocarta MAPK pathways | 31 | 2.802 |
| REACTOME | Interaction between L1 and ankyrins | 13 | 2.585 |
| REACTOME | Fatty acid triacylglycerol and ketone body metabolism | 22 | 2.563 |
| KEGG | WNT signaling pathway | 36 | 2.559 |
| REACTOME | Neuronal system | 59 | 2.365 |
| PATHWAY INTERACTION DATABASE | Neurotrophic factor-mediated Trk receptor signaling | 16 | 2.331 |
| KEGG | Focal adhesion | 48 | 2.328 |
| KEGG | Glioma | 20 | 2.323 |
| KEGG | Chronic myeloid leukemia | 20 | 2.307 |
| KEGG | Neurotrophin signaling pathway | 29 | 2.241 |
| PATHWAY INTERACTION DATABASE | C-myb transcription factor network | 26 | 2.237 |
| REACTOME | Signaling by NGF | 49 | 2.194 |
| KEGG | Pancreatic cancer | 21 | 2.138 |
| KEGG | TGF-beta signaling pathway | 28 | 2.129 |
| PATHWAY INTERACTION DATABASE | Signaling events regulated by Ret tyrosine kinase | 11 | 2.103 |
| REACTOME | NGF signaling via TrkA from Plasma membrane | 37 | 2.057 |
| Database | Pathway | Genes | Raw | Empirical |
|---|---|---|---|---|
| REACTOME | Signaling by insulin receptor | 4 | 1.41372e-2 | 5.07712e-2 |
| KEGG | Focal adhesion | 6 | 7.34030e-3 | 3.22732e-2 |
| KEGG | MAPK signaling pathway | 7 | 8.68277e-3 | 8.62286e-2 |
| KEGG | Neurotrophin signaling pathway | 6 | 2.23557e-2 | 1.32203e-1 |
| REACTOME | Signaling to ERKs | 3 | 3.57835e-3 | 3.10591e-2 |
| KEGG | Pathways in cancer | 5 | 9.69353e-2 | 3.33703e-1 |
| REACTOME | Signaling by NGF | 6 | 1.14105e-2 | 5.61302e-2 |
| REACTOME | NGF signaling via TrkA from plasma membrane | 5 | 6.46094e-3 | 1.29899e-2 |
| REACTOME | Hemostasis | 5 | 1.66621e-1 | 3.67254e-1 |
| Database | Pathway | Genes | Raw | Empirical |
|---|---|---|---|---|
| REACTOME | Developmental Biology | 494 | 8.78e-4 | 3.89749e-1 |
| REACTOME | Adaptive immune system | 482 | 2.519e-2 | 2.90419e-1 |
| REACTOME | Hemostasis | 467 | 6.955e-3 | 3.12984e-1 |
| REACTOME | Transmembrane transport of small molecules | 427 | 1.07902e-1 | 5.3854e-1 |
| REACTOME | GPCR ligand binding | 410 | 1.5103e-2 | 5.7303e-2 |
| REACTOME | Cell cycle mitotic | 330 | 1.06856e-1 | 4.15975e-1 |
| KEGG | Pathways in cancer | 325 | 3.1018e-2 | 5.8577e-1 |
| KEGG | Neuroactive ligand receptor activation | 318 | 9.2561e-2 | 2.12222e-1 |
| REACTOME | Class A1 (Rhodopsin-Like receptors) | 305 | 6.7227e-2 | 2.74648e-1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Putkonen, N.; Laiho, A.; Ethell, D.; Pursiheimo, J.; Anttonen, A.-K.; Pitkonen, J.; Gentile, A.M.; de Diego-Otero, Y.; Castrén, M.L. Urine microRNA Profiling Displays miR-125a Dysregulation in Children with Fragile X Syndrome. Cells 2020, 9, 289. https://doi.org/10.3390/cells9020289
Putkonen N, Laiho A, Ethell D, Pursiheimo J, Anttonen A-K, Pitkonen J, Gentile AM, de Diego-Otero Y, Castrén ML. Urine microRNA Profiling Displays miR-125a Dysregulation in Children with Fragile X Syndrome. Cells. 2020; 9(2):289. https://doi.org/10.3390/cells9020289
Chicago/Turabian StylePutkonen, Noora, Asta Laiho, Doug Ethell, Juha Pursiheimo, Anna-Kaisa Anttonen, Juho Pitkonen, Adriana M. Gentile, Yolanda de Diego-Otero, and Maija L. Castrén. 2020. "Urine microRNA Profiling Displays miR-125a Dysregulation in Children with Fragile X Syndrome" Cells 9, no. 2: 289. https://doi.org/10.3390/cells9020289
APA StylePutkonen, N., Laiho, A., Ethell, D., Pursiheimo, J., Anttonen, A. -K., Pitkonen, J., Gentile, A. M., de Diego-Otero, Y., & Castrén, M. L. (2020). Urine microRNA Profiling Displays miR-125a Dysregulation in Children with Fragile X Syndrome. Cells, 9(2), 289. https://doi.org/10.3390/cells9020289