1. Introduction
Prostate cancer (PCa) represents the most frequently diagnosed cancer in men in the developed countries of Europe and USA [
1,
2]. Although radical surgery and radiotherapy turned out to be effective to control localized PCa, a significant proportion of patients still experience relapse, in the form of biochemical or local recurrence or metastasis [
3]. The efficacy of androgen-deprivation therapy, the treatment of choice for metastatic tumors, is itself limited in time, thus making the management of castration-resistant disease challenging and the patient’s prognosis invariably poor [
4]. In this scenario, the development of novel therapies or the implementation of currently used approaches is an urgent need.
MicroRNAs (miRNAs), endogenous small noncoding RNAs that negatively regulate gene expression at the post-transcriptional level [
5], have been found aberrantly expressed in several human diseases, including cancer [
6,
7]. Specifically, depending on their target genes, miRNAs have been acknowledged to exert either pro- or anti-metastatic functions [
8].
Based on the available literature, it is still difficult to outline the precise signature of miRNAs related to PCa pathogenesis and progression, however several miRNAs are emerging as master regulators of processes involved in different steps of cancer evolution and in drug response. For example,
miR-29b, the expression of which is lower in PCa cells compared to normal cells, has been recently reported to inhibit prostate tumor growth in vivo and induce apoptosis by enhancing Bim expression [
9]. Down-regulation of
miR-130b in PCa cells was instead shown to significantly promote the proliferation, invasion and tubule formation of human umbilical vein endothelial cells, while ectopic expression of
miR-130b blocked prostate cancer angiogenesis in vitro and in vivo [
10]. We demonstrated that
miR-205, a down-regulated miRNA in PCa tissues, is able to pleiotropically hamper PCa metastasis by reverting epithelial to mesenchymal transition (EMT) [
11] and restoring the deposition of basement membrane by tumor cells [
12]. Consistent with the role of EMT in the establishment of radioresistance [
13],
miR-205 can also increase response of PCa cells to ionizing radiation [
14]. A tumor-suppressive behavior comparable to that of
miR-205 was reported for
miR-875-5p, which controls a different set of target genes [
15].
Several efforts have been and are still being made to establish miRNA involvement in PCa with the long term perspective to introduce miRNA-based approaches as a novel strategy to withstand the disease, alone or in combination with the currently used therapies. Since the prelude to the development of miRNA-based therapies is the analysis of miRNA function in PCa initiation and progression, in this study, for the first time, we provide evidence of a putative tumor-suppressive role of miR-1272.
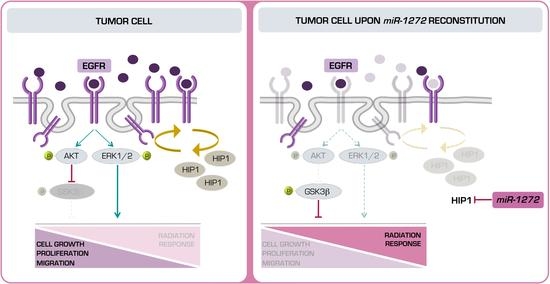
Specifically, we found that miR-1272 is down-modulated in PCa samples with respect to normal counterparts. In addition, we showed that, when restored in a couple of metastatic PCa cell lines, miR-1272 is able to hinder EMT, drastically reduce migration and invasion, limit cell growth and act as radiosensitizer by reducing the levels of Huntingtin Interacting Protein 1 (HIP1), whose overexpression has been associated with PCa and correlated with the severity of the disease.
2. Materials and Methods
2.1. Cell Culture
Established human PCa cell lines were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA) and cultured in standard conditions. DU145 and 22Rv1 cells were cultured in RPMI-1640 medium (Lonza, Basel, Switzerland) supplemented with 10% FBS (Thermo Fisher Scientific Inc., Waltham, MA, USA). Cell lines were authenticated and periodically monitored by genetic profiling using short tandem repeat analysis (AmpFISTR Identifiler PCR amplification kit, Thermo Fisher Scientific Inc., Waltham, MA, USA). Cells were routinely checked for possible mycoplasma contamination through MycoAlert® Mycoplasma Detection Kit (Lonza, Basel, Switzerland).
Cell morphology was evaluated usually at day 3 after transfection using an Eclipse TE2000-S microscope (Nikon, Japan). Images were acquired by a Digital Camera DXM100F (Nikon, Japan).
2.2. Transfection
Cells were seeded at the density of 8000 cells/cm2 in culture vessels. Twenty-four hours later, medium was removed and cells were transfected with 20 nM mirVana miRNA mimic (miR-1272 MC13413, Thermo Fisher Scientific Inc., Waltham, MA, USA) or 30 nM siRNA (siHIP1, in sense format 5′-GGCUUAGGAUCGACAAGAA-3′, purchased from Eurofins MWG-Biotech, Ebesberg, Germany) for 4 h, using Optimem medium (Thermo Fisher Scientific Inc., Waltham, MA, USA) and Lipofectamine-2000 reagent (Thermo Fisher Scientific Inc., Waltham, MA, USA) according to the manufacturer’s protocol. A control mirVana miRNA mimic (miR-Neg mirVanaTM miRNA mimic Negative control #1, Thermo Fisher Scientific Inc., Waltham, MA, USA) and a control siRNA (siCtr, in sense format 5′-GCAUACAAUGGAGUUGUUA-3′, purchased from Eurofins MWG-Biotech, Ebesberg, Germany) with no homology to any known human mRNA were used for comparative analyses.
2.3. Cell Growth Evaluation
Cells were seeded in 6-well plates at the density of 4000 cells/cm
2 and transfected with miRNA mimics or siRNAs 24 h later, as previously described. Daily, from day one to five, cells were detached and counted through a Coulter Counter (Beckman Coulter Life Sciences, Brea, CA, USA). Data were reported as cell percentage with respect to the number of
miR-Neg or
siCtr cells collected at day 1 (24 h). Cell doubling time of each cell line was calculated from growth curves of parental cells, as described in [
16]. Staining for Ki-67 was determined by immunohistochemistry. Briefly, transfected cells were removed from dishes through scraper, formalix-fixed and paraffin-embedded. Some sections were then deparaffinised in xylene, rehydrated through graded alcohols to water, and subjected to immunohistochemical analysis using Ki-67 antibody (MIB-1, Dako; 1:200). Nuclei were counterstained with hematoxylin. Images were acquired by Nikon Eclipse E600 microscope using ACT-1 software (Nikon). At least 10 fields were scanned and the average number of Ki-67-positive and negative cells was plotted.
2.4. Apoptosis Analysis
Cell apoptosis was evaluated in terms of catalytic activity of Caspase-3 by using the APOPCYTO Caspase-3 Colorimetric Assay Kit (MBL International Corporation, Woburn, MA, USA), according to manufacturer’s protocol. Briefly, at 96 h after transfection, cells were detached, lysed and extracted proteins were incubated with the substrate N-acetylAsp-Glu-Val-Asp-AMC (DEVD-AMC). The hydrolysis of the proper substrate was evaluated through spectrofluorometry with 380-nm excitation and 460-nm emission filters by using POLARstar OPTIMA plate reader (BMG Labtech, Ortenberg, Germany). For terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay, transfected cells were fixed and treated by using the In Situ Cell Death Detection Kit (Roche) according to manufacturer’s instructions. The cells were subjected to FACS analysis (BD Accuri™ C6 Cytometer, Becton Dickinson, Basel CH) and data were reported in graph as the percentage of positive cells.
2.5. Migration and Invasion Assays
For migration and invasion assays, cells were cultured and transfected for 72 h as previously described and starved in serum-free medium for 24 h. Cells were transferred to the upper chamber of 24-well Transwell plates (Costar, Corning Incorporated, New York, NY, USA) in serum-free medium at a concentration of 120,000 cells/well. Medium supplemented with 10% of FBS was added to the lower chamber. After a 6 h-incubation at 37 °C, filters were fixed in 99% ethanol and stained with a 0.4% sulforhodamine B/1% acetic acid solution. Migrated cells were counted under a microscope. The same procedure was used for invasion assay, except that cells were seeded at 240,000 cells/well, Transwell chambers coated with 12.5 μg of Matrigel/well (BD Biosciences, San Jose, CA, USA), and samples processed after a 24 h-incubation.
2.6. Total RNA Extraction and RT-qPCR
RNA extraction and cDNA synthesis were performed as detailed in [
17]. Briefly, RNA was isolated using QIAzol Lysis Reagent and miRNeasy Mini Kit (QIAGEN, Hilden, Germany) with DNase I digestion (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. cDNA was synthesized using High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific Inc., Waltham, MA, USA). For miRNA detection, TaqMan MicroRNA Reverse Transcription kit and sequence specific primers (Thermo Fisher Scientific Inc., Waltham, MA, USA) were used.
Quantification of gene or miRNA expression was assessed by RT-qPCR as widely described in [
16] using No AmpErase TaqMan Universal PCR Master Mix (Thermo Fisher Scientific Inc., Waltham, MA, USA) and the specific TaqMan gene-expression assays (Thermo Fisher Scientific Inc., Waltham, MA, USA), as follows:
miR-1272 (TaqMan
® microRNA assay PN4427975),
CDH1 (TaqMan
® gene expression assay Hs00170423_m1),
CTNNB1 (TaqMan
® gene expression assay Hs00355045_m1),
VIM (TaqMan
® gene expression assay Hs00185584_m1). For comparative analyses,
GAPDH (PN4326317E) and
RNU48 (TaqMan
® microRNA assay PN4440887) were used as endogenous controls for genes and miRNA, respectively. RT-qPCR results were reported as −ddCt or relative quantity (RQ = 2
−ddCt) with respect to a calibrator sample using the comparative Ct (ddCt) method.
2.7. Gene Expression and Bioinformatic Analyses
Transcriptomic profile of DU145 cells transfected with miR-1272 synthetic mimic and those transfected with mock molecule (miR-Neg) were collected in triplicate from scanned images using Illumina BeadStudio v3.3.8 (Illumina) and processed using the lumi package [
18] from Bioconductor v3.0 [
19]. Raw data were log
2-transformed, normalized with Robust Spline Normalization and filtered, keeping only probes with a detection
p-value < 0.01 in at least one sample and probes not associated to official gene symbol were removed. Expression data were deposited in the Gene Expression Omnibus repository (GEO) with accession number GSE68883.
Differentially expressed genes between the two conditions were identified using the
limma package [
20], and significance was assessed by Benjamini–Hochberg false discovery rate (FDR) method in order to take into account the multiple-testing correction. Genes showing an FDR < 0.05 were considered significantly differentially expressed. Selecting only significantly up (fold change, FC > 0) and down regulated (FC < 0) genes an overrepresentation analysis was performed using WebGestalt on-line available tool [
21]. Only enrichment with an FDR above 0.05 was considered significant.
miRNA expression data of tumor and non-neoplastic prostate samples from PCa patients were retrieved from GEO repository with accession number GSE76260, in the form of normalized data matrix.
2.8. In Silico miR-1272 Target Prediction
Prediction of
miR-1272 target genes was accomplished using miRWalk (
http://mirwalk.umm.uni-heidelberg.de/) and MtiBase online tools. The former solely contains in silico predictions from several target-prediction databases while the latter also includes information from experimental data such as ultraviolet cross-linking and immunoprecipitation (CLIP), microarray and RNA sequencing, ribosome-protected fragment sequencing and pulse stable isotope labelling with amino acids in culture (pSILAC) [
22]. For miRWalk analysis, target genes were selected as those predicted by at least one database, while for MtiBase at least two prediction databases were considered in association with at least three supporting experiments.
2.9. Protein Extraction, Cytoplasmic/Membrane Fractionation and Western Blotting
Total protein extraction and immunoblotting experiments were performed as fully detailed in [
16]. In this work the following antibodies were used: AKT (cs9272, Cell Signaling Technologies
®, Danvers, MA, USA; 1:1000), β-catenin (sc59891, Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:200), β-tubulin (ab6160, abcam
®, Cambridge, UK; 1:500), Caveolin-1 (610406, BD Biosciences, San Jose, CA, USA; 1:500), Cleaved PARP (cs9541, Cell Signaling Technologies
®, Danvers, MA, USA; 1:1000), E-cadherin (sc7870, Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:500), EGFR (sc03, Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:200), ERK1/2 (ab17942, abcam
®, Cambridge, UK; 1:1000), GSK3β (sc9166, Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:200), HIP1 (MA1-16747, Thermo Fisher Scientific Inc., Waltham, MA, USA; 1:500), PARP (cs9542, Cell Signaling Technologies
®, Danvers, MA, USA; 1:1000), p-AKT (ab27773, abcam
®, Cambridge, UK; 1:500), p-ERK1/2 (cs9101, Cell Signaling Technologies
®, Danvers, MA, USA; 1:1000), p-GSK3β (S9, R&D System, Minneapolis, MN, USA; 1:1000), Slug (cs9585, Cell Signaling Technologies
®, Danvers, MA, USA; 1:1000), Vimentin (sc32322, Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:200), Vinculin (V9131, Sigma-Aldrich, St. Louis, MO, USA; 1:5000).
For cytoplasmic/membrane protein separation, the Subcellular Proteins Fractionation Kit for Cultured Cells (Thermo Fisher Scientific Inc., Waltham, MA, USA) was used according to manufacturer’s protocol. Briefly, cultured cells were harvested, treated with buffers to selectively fractionate proteins from cytoplasm (cytoplasmic extraction buffer) and membrane (membrane extraction buffer), and properly centrifuged to enhance protein separation. Immunoblotting experiments were performed as for total proteins.
2.10. Immunofluorescence
Transfected cells grown for 72 h on μ-Slides 8 Well coverlips (ibidi®) were fixed with 4% formaldehyde and permeabilised with cold 70% ethanol. Cells were probed for 1 h with primary antibodies as follows: β-catenin (ab2982, abcam®, Cambridge, UK), β-tubulin (ab6046, abcam®, Cambridge, UK) and E-cadherin (ab1416, abcam®, Cambridge, UK); and subsequently with Alexa Fluor448-labeled or Alexa Fluor594-labeled secondary antibodies (Thermo Fisher Scientific Inc., Waltham, MA, USA) for 1 h at room temperature. Nuclei were counterstained with DAPI (ab104139, abcam®, Cambridge, UK; 1:15,000). Images were acquired by Nikon Eclipse E600 microscope using ACT-1 software (Nikon) and processed with ImageJ.
2.11. In Vivo Experiments
Experimental protocols were approved by the Ethics Committee for Animal Experimentation of Fondazione IRCCS Istituto Nazionale dei Tumori, and by the Italian Healthy Ministry (project identification code: 1120/2015-PR. Date of approval: 22nd October 2015). Ten million miR-Neg or miR-1272 DU145 cells were subcutaneously injected into the right flank of eight-week-old male SCID mice (Charles River, Calco, Italy). Each group contained ten mice. Animals were maintained under standard light and temperature conditions and had free access to food and water. Tumor growth was determined by measurement of tumor size with a Vernier caliper.
2.12. Irradiation Experiments
Irradiation experiments on DU145 cells transfected with
miR-Neg or
miR-1272 were performed as in El Bezawy et al. [
14]. At day 10 after irradiation, colonies were fixed, stained and counted. The colony-forming efficiency was calculated as the ratio of the number of colonies (consisting of at least 50 cells) to the number of single cells seeded. The surviving fraction was calculated as the ratio of the colony-forming efficiency of the irradiated sample to that of the non-irradiated one. Triplicate wells were set up for each condition. Data are reported as a linear-quadratic equation, which has been well established to be a suitable model for the description of cell death upon irradiation [
23].
2.13. Statistical Analyses
If not otherwise specified, data are presented as mean values +/± SD from at least three independent experiments. Statistical analysis was performed by two-tailed Student’s t test except for in vivo experiment where Mann–Whitney U test was applied. p-values < 0.05 were considered statistically significant.
4. Discussion
Compelling evidence about the involvement of endocytic proteins in cancer initiation and progression has emerged recently [
26]. However, the exact mechanism by which a disrupted regulation of the endocytic pathway may lead a normal cell to become cancerous is still completely unclear [
26]. Several proteins are involved in endocytosis/clathrin-mediated pathways and in turn copious membrane-protein (e.g., growth factor receptors) turn-over is finely regulated by these mechanisms. In this regard, an impaired/unbalanced receptor-trafficking mediated by endocytosis has been associated to cell transformation and cancer [
26].
Chiefly, HIP1, a cofactor in clathrin-mediated vescicle trafficking, was firstly found to be implicated in cancer as part of a chromosomal translocation in patients with chronic myelomonocytic leukemia [
28]. Other studies revealed HIP1 to be deregulated in several human solid tumors [
26]. Particularly, HIP1 expression was demonstrated to be more elevated in prostate and colon cancers with respect to their normal counterparts. In addition, HIP1 expression increased in parallel with PCa progression up to metastasis. Consistent with this evidence, primary prostate tumors lacking HIP1 expression were characterized by reduced frequency of disease progression, in terms of biochemical relapse, after radical prostatectomy [
26]. Altogether, these findings suggested HIP1 as a putative prognostic biomarker and therapeutic target for PCa management [
26].
Different layers of regulation may impact on HIP1 expression. In the last decades, miRNAs have gained increasing respect as regulators of various biological processes [
29]. Hence, aberrant miRNA expression has been causatively associated with the pathogenesis of several diseases, including cancer [
7]. Depending on the cellular context and the functions of their targets, miRNAs may act as oncomirs or tumor-suppressors. Generally, the down-modulation of a tumor-suppressor miRNA capable of controlling the expression of an oncogene may result in deregulated over-expression of the target, thus contributing to tumor development and progression [
30]. Several tumor-suppressive miRNAs have been identified in PCa context, and their role clarified through functional studies. However, the function of
miR-1272 in PCa, and in human cancer in general, has not been explored so far. Here, we show that
miR-1272 is downregulated in PCa compared to adjacent normal tissue. The reconstitution of
miR-1272 expression in PCa cells, inherently expressing negligible levels of the miRNA, resulted in a marked abrogation of its predicted target
HIP1. Notably, the results of 6 Ago-CLIP sequencing experiments collected in Mtibase (
Table 1) confirmed that
miR-1272 binding site within
HIP1 3′UTR is not merely predicted in silico but is also physically bound by Ago proteins within RNA-induced silencing complex. Such evidence confirms the functional repression of
HIP1 by
miR-1272, though not relying on the conventional luciferase reporter assays. Our results pave the way for future studies aimed at verifying whether pathological loss of
miR-1272 in normal cells may lead to HIP1 over-expression, which in turn may dysregulate the endocytic pathway, ultimately disrupting signalling pathways that control growth, survival and migration.
Aberrant endocytic turnover may lead to the amplification of signal transduction cascades mediated by a sustained activation of growth factor receptors (e.g., EGFR). The upregulation of EGFR signalling has been widely associated with more aggressive behaviour in several human cancers [
31]. In addition, in the context of PCa, the acquisition of a hormone-refractory state was suggested to be related to an excessive activation of the EGFR axis and the downstream pathways, including PI3K-AKT and ERK, thus impinging on several tumor traits [
32]. In our PCa models,
miR-1272 supplementation significantly reduced EGFR membrane exposition, presumably as a consequence of the restored correct receptor turn-over. Interestingly, such phenotype was recapitulated by the direct silencing of
HIP1.
MiR-1272-mediated impairment of EGFR signalling resulted in reduced activation of ERK1/2 and AKT-GSK3β axes, which ultimately led to reduced cell growth and migration, respectively. Similar results were reported in the context of fibroblast-like synoviocyte models of rheumatoid arthritis, one of the most common autoimmune diseases, where HIP1 expression was shown to be required for receptor tyrosine kinase (RTK) stabilization on the cell surface. In this regard, HIP1 abrogation was able to reduce fibroblast-like synoviocyte invasion proficiency by interfering with lamellipodia formation and cytoskeletal re-organization, a phenotype sustained by the deregulation of ERK1/2 signalling upon HIP1 silencing [
33]. Again, during angiogenesis (
i.e., the formation of new blood vessels from the pre-existing ones) for tissue repair in various pathological conditions such as ischemic cardiovascular events, endothelial cells are stimulated by VEGF to proliferate and migrate. In this context,
miR-135a-3p was shown to be able to impair p38 signalling in response to VEGF stimulation through the suppression of HIP1. In addition, endothelial cell growth and migration resulted severely affected upon
miR-135a-3p restoration [
34]. Hence, the disruption of membrane receptor turn-over mediated by HIP1 shows comparable phenotypes and involves similar pathways in completely different settings. In the context of Huntington Disease (HD), another epigenetic regulator of
HIP1 and of other HD-related factors is
miR-128a [
35]. The same miRNA was identified upon an integrative analysis of proteomic changes during PCa progression. The proteomic signature revealed the involvement of
miR-128a (and
miR-128b) in regulating the transition from organ-confined disease to metastasis. Precisely,
miR-128 levels were reduced in invasive PCa compared to benign prostate epithelial, and
miR-128 overexpression was able to attenuate invasion of PCa cells [
36]. Comprehensively, miRNAs capable (or supposed) to target
HIP1 were demonstrated to impair/affect migration and invasion, the prelude of which (in epithelial tumors) is EMT.
In this study, we elucidated the role of
miR-1272 as repressor of EMT process. Aside from being a crucial step for the metastatic dissemination, which represents the main cause of death in PCa patients, EMT was reported as related to radioresistance in several tumors [
37]. In PCa context, radiation therapy represents a standard of care whereas radiation resistance remains the main obstacle for long-term treatment efficacy. In fact, upon radiation, cancer cells activate several different pathways to overcome cytotoxic effects of treatment. In breast cancer, ionizing radiation was found to enable HER RTKs and the downstream pathways such as AKT and ERK1/2 [
38]. The pro-survival effects mediated by the aforementioned pathways are related to the abolishment of the apoptotic stimuli and, concurrently, to the induction of cell cycle checkpoint response and DNA repair. In this regard, similar mechanisms of miRNA-mediated radiosensitization through RTKs have been recently described in PCa models. Primarily,
miR-205, the role of which elicits great interest in PCa, was demonstrated to be involved in regulating EGFR nuclear-translocation, a mechanism known to control ionizing radiation response, by targeting PKCε [
14]. In addition, in both prostate and breast cancers,
miR-205 revealed to be capable to enhance radiation response by targeting ZEB1, a well-established EMT inhibitor [
14,
15,
16,
17,
18,
19,
20,
21,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31,
32,
33,
34,
35,
36,
37,
38,
39]. Similarly, another miRNA,
miR-875-5p, was found to radiosensitize PCa cells by targeting EGFR and thereby reducing receptor nuclear-translocation [
15]. Altogether, these findings suggest that the complex network existing between miRNAs, EGFR, ERK1/2 axis and EMT process is involved in radioresistance. Here, we proved, for the first time, the radiosensitizing effect of
miR-1272 in PCa, and showed that this effect is at least in part mediated by
HIP1 suppression.
Overall, a putative therapeutic approach based on miR-1272 reconstitution in association with ionizing radiation may improve PCa response to conventional radiotherapy, as we showed that miR-1272 can reduce the activity of EGFR/AKT/ERK1-2 pathways, known to be triggered as pro-survival signals upon irradiation. A potential clinical interest for miR-1272 may also lie on its ability to affect cell growth and EMT. In this regard, a miR-1272-based therapy in an organ-confined PCa context may primarily represent a suitable strategy to reduce tumor growth and enhance response to radiotherapy. Additionally, miR-1272 may also prevent EMT, the first step of metastatic cascade. Prerequisite to the development of any miR-1272-based therapeutic approach would be the setting up of miRNA mimic delivery strategies streamlined for efficiency and specificity of prostate targeting.