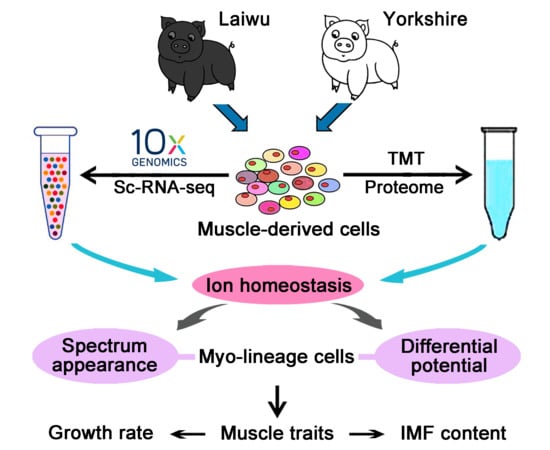
Association Analysis of Single-Cell RNA Sequencing and Proteomics Reveals a Vital Role of Ca2+ Signaling in the Determination of Skeletal Muscle Development Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Samples
2.3. Preparation of Muscle-Derived Cell Suspension
2.4. Primary Cell Isolation, Culture, and Differentiation
2.5. Single-Cell RNA Sequencing
2.6. Tandem Mass Tag (TMT) Labeling and LC-MS/MS Analysis
2.7. Protein Identification and Bioinformatics Analysis
2.8. Quantitative Real-Time PCR Analysis
2.9. Western Blot
2.10. Immunocytochemistry
2.11. Determination of Cellular Ion Concentration
2.12. Statistical Analysis
3. Results
3.1. The Differentiation Potential of Muscle-Derived Cells
3.2. Profiling and Trajectory Analysis of Myo-Lineage Cells
3.3. Quantitative Mapping of the Proteome of Myo-Lineage Cells
3.4. Bioinformatics Exploration of DEPs Via GO and KEGG Analyses
3.5. Distinct Cellular Ion Regulation between Myo-L and Myo-Y
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Goodpaster, B.H.; Sparks, L.M. Metabolic Flexibility in Health and Disease. Cell Metab. 2017, 25, 1027–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building muscle: Molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008342. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft, A.J.; Sayer, A.A. Sarcopenia. Lancet 2019, 393, 2636–2646. [Google Scholar] [CrossRef]
- Chang, N.C.; Chevalier, F.P.; Rudnicki, M.A. Satellite Cells in Muscular Dystrophy—Lost in Polarity. Trends Mol. Med. 2016, 22, 479–496. [Google Scholar] [CrossRef] [Green Version]
- Katsanos, C.S.; Mandarino, L.J. Protein metabolism in human obesity: A shift in focus from whole-body to skeletal muscle. Obesity 2011, 19, 469–475. [Google Scholar] [CrossRef]
- Hilton, T.N.; Tuttle, L.J.; Bohnert, K.L.; Mueller, M.J.; Sinacore, D.R. Excessive adipose tissue infiltration in skeletal muscle in individuals with obesity, diabetes mellitus, and peripheral neuropathy: Association with performance and function. Phys. Ther. 2008, 88, 1336–1344. [Google Scholar] [CrossRef] [Green Version]
- Tachi, Y.; Kozuka, A.; Hirai, T.; Kojima, Y.; Ishizu, Y.; Honda, T.; Kuzuya, T.; Hayashi, K.; Ishigami, M.; Goto, H. Skeletal muscle fat deposition is associated with hepatocellular carcinoma development in patients with chronic liver disease. Nutrition 2018, 54, 83–88. [Google Scholar] [CrossRef]
- Robles, P.G.; Sussman, M.S.; Naraghi, A.; Brooks, D.; Goldstein, R.S.; White, L.M.; Mathur, S. Intramuscular Fat Infiltration Contributes to Impaired Muscle Function in COPD. Med. Sci. Sport. Exerc. 2015, 47, 1334–1341. [Google Scholar] [CrossRef]
- Gandemer, G. Lipids in muscles and adipose tissues, changes during processing and sensory properties of meat products. Meat Sci. 2002, 62, 309–321. [Google Scholar] [CrossRef]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef] [Green Version]
- Pagano, A.F.; Brioche, T.; Arc-Chagnaud, C.; Demangel, R.; Chopard, A.; Py, G. Short-term disuse promotes fatty acid infiltration into skeletal muscle. J. Cachexia Sarcopenia Muscle 2018, 9, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Park, K.W.; Halperin, D.S.; Tontonoz, P. Before they were fat: Adipocyte progenitors. Cell Metab. 2008, 8, 454–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porpiglia, E.; Samusik, N.; Ho, A.T.V.; Cosgrove, B.D.; Mai, T.; Davis, K.L.; Jager, A.; Nolan, G.P.; Bendall, S.C.; Fantl, W.J.; et al. High-resolution myogenic lineage mapping by single-cell mass cytometry. Nat. Cell Biol. 2017, 19, 558–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delfini, M.C.; Hirsinger, E.; Pourquié, O.; Duprez, D. Delta 1-activated notch inhibits muscle differentiation without affecting Myf5 and Pax3 expression in chick limb myogenesis. Development 2000, 127, 5213–5224. [Google Scholar] [PubMed]
- Achim, K.; Arendt, D. Structural evolution of cell types by step-wise assembly of cellular modules. Curr. Opin. Genet. Dev. 2014, 27, 102–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, J.; Spielmann, M.; Qiu, X.; Huang, X.; Ibrahim, D.M.; Hill, A.J.; Zhang, F.; Mundlos, S.; Christiansen, L.; Steemers, F.J.; et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496–502. [Google Scholar] [CrossRef]
- Perleberg, C.; Kind, A.; Schnieke, A. Genetically engineered pigs as models for human disease. Dis. Model. Mech. 2018, 11, dmm030783. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; Irie, M.; Kadowaki, H.; Shibata, T.; Kumagai, M.; Nishida, A. Genetic parameter estimates of meat quality traits in Duroc pigs selected for average daily gain, longissimus muscle area, backfat thickness, and intramuscular fat content. J. Anim. Sci. 2005, 83, 2058–2065. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Wang, H.; Jie, L.; Jie, W.; Zheng, M.; Tan, X.; Xing, S.; Cui, H.; Li, Q.; Zhao, G. Uncovering the embryonic development-related proteome and metabolome signatures in breast muscle and intramuscular fat of fast-and slow-growing chickens. BMC Genom. 2017, 18, 816. [Google Scholar] [CrossRef]
- Qiu, K.; Zhang, X.; Wang, L.; Jiao, N.; Xu, D.; Yin, J. Protein expression landscape defines the differentiation potential specificity of adipogenic and myogenic precursors in the skeletal muscle. J. Proteome Res. 2018, 17, 3853–3865. [Google Scholar] [CrossRef]
- Zheng, G.X.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, N.J.; Church, C.D.; Dong, L.; Crispin, D.; Fitzgibbon, M.P.; Lachance, K.; Jing, L.; Shinohara, M.; Gavvovidis, I.; Willimsky, G.; et al. Tumor-infiltrating merkel cell polyomavirus-specific T cells are diverse and associated with improved patient survival. Cancer Immunol. Res. 2017, 5, 137–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Huang, Y.; Das, A.K.; Yang, Q.; Duarte, M.S.; Dodson, M.V.; Zhu, M.J. Meat science and muscle biology symposium: Manipulating mesenchymal progenitor cell differentiation to optimize performance and carcass value of beef cattle. J. Anim. Sci. 2013, 91, 1419–1427. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Kim, J.R.; Moon, J.K.; Choi, B.H.; Kim, T.H.; Kim, K.S.; Kim, J.J.; Lee, C.K. Transcriptional alteration of p53 related processes as a key factor for skeletal muscle characteristics in Sus scrofa. Mol. Cells 2009, 28, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, Y.; Hou, L.; Ma, J.; Chen, C.; Ai, H.; Huang, L.; Ren, J. Genome-wide detection of genetic markers associated with growth and fatness in four pig populations using four approaches. Genet. Sel. Evol. 2017, 49, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briggs, J.A.; Weinreb, C.; Wagner, D.E.; Megason, S.; Peshkin, L.; Kirschner, M.W.; Klein, A.M. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 2018, 360, eaar5780. [Google Scholar] [CrossRef] [Green Version]
- Dell’Orso, S.; Juan, A.H.; Ko, K.D.; Naz, F.; Perovanovic, J.; Gutierrez-Cruz, G.; Feng, X.; Sartorelli, V. Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions. Development 2019, 146, dev181743. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Michowski, W.; Kolodziejczyk, A.; Sicinski, P. The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat. Cell Biol. 2019, 21, 1060–1067. [Google Scholar] [CrossRef]
- Zhu, C.C.; Dyer, M.A.; Uchikawa, M.; Kondoh, H.; Lagutin, O.V.; Oliver, G. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development 2002, 129, 2835–2849. [Google Scholar]
- Seale, P.; Sabourin, L.A.; Girgis-Gabardo, A.; Mansouri, A.; Gruss, P.; Rudnicki, M.A. Pax7 is required for the specification of myogenic satellite cells. Cell 2000, 102, 777–786. [Google Scholar] [CrossRef] [Green Version]
- Kassar-Duchossoy, L.; Gayraud-Morel, B.; Gomès, D.; Rocancourt, D.; Buckingham, M.; Shinin, V.; Tajbakhsh, S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 2004, 431, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, N.; Papadakis, M.; Wilson, M.; Malhotra, A.; Adamuz, C.; Homfray, T.; Monserrat, L.; Behr, E.R.; Sharma, S. Diagnostic yield of genetic testing in young athletes with T-wave inversion. Circulation 2018, 138, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, L.; Qiu, K.; Xu, D.; Yin, J. Dynamic membrane proteome of adipogenic and myogenic precursors in skeletal muscle highlights EPHA2 may promote myogenic differentiation through ERK signaling. FASEB J. 2019, 33, 5495–5509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; He, T.; Qin, C.; Qiu, K.; Zhang, X.; Luo, Y.; Li, D.; Yin, J. A potential regulatory network underlying distinct fate commitment of myogenic and adipogenic cells in skeletal muscle. Sci. Rep. 2017, 7, 44133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernst, C.W.; Mcfarland, D.C.; White, M.E. Expression of insulin-like growth factor II (IGF-II), IGF binding protein-2 and myogenin during differentiation of myogenic satellite cells derived from the turkey. Differentiation 1996, 61, 25–33. [Google Scholar] [CrossRef]
- Abdellatif, A.; Zubasurma, E.K.; Case, J.; Tiwari, S.; Hunt, G.; Ranjan, S.; Vincent, R.J.; Srour, E.F.; Bolli, R.; Dawn, B. TGF-β1 enhances cardiomyogenic differentiation of skeletal muscle-derived adult primitive cells. Basic Res. Cardiol. 2008, 103, 514–524. [Google Scholar] [CrossRef] [Green Version]
- Jr, M.R.; Nindl, B.C. Loss of IGF-IEa or IGF-IEb impairs myogenic differentiation. Endocrinology 2011, 152, 1923–1934. [Google Scholar]
- Brodsky, F.M.; Chen, C.Y.; Knuehl, C.; Towler, M.C.; Wakeham, D.E. Biological Basket Weaving: Formation and Function of Clathrin-Coated Vesicles. Annu. Rev. Cell Dev. Biol. 2001, 17, 517–567. [Google Scholar] [CrossRef] [Green Version]
- Wakeham, D.E.; Abi-Rached, L.; Towler, M.C.; Wilbur, J.D.; Parham, P.; Brodsky, F.M. Clathrin heavy and light chain isoforms originated by independent mechanisms of gene duplication during chordate evolution. Proc. Natl. Acad. Sci. USA 2005, 102, 7209–7214. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.V. Dual roles of palladin protein in in vitro myogenesis: Inhibition of early induction but promotion of myotube maturation. PLoS ONE 2015, 10, e0124762. [Google Scholar]
- Wanlong, H.; Xiuxiu, Z.; Ai, L.; Xiangyang, M. Identification of differentially expressed genes between subcutaneous and intramuscular adipose tissue of Large White pig using RNA-seq. Hereditas 2017, 39, 501–511. [Google Scholar]
- Sun, L.; Yu, Y.; Huang, T.; An, P.; Yu, D.; Yu, Z.; Li, H.; Sheng, H.; Cai, L.; Xue, J.; et al. Associations between ionomic profile and metabolic abnormalities in human population. PLoS ONE 2012, 7, e38845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otero-Rey, E.M.; Somoza-Martín, M.; Barros-Angueira, F.; García-García, A. Intracellular pH regulation in oral squamous cell carcinoma is mediated by increased V-ATPase activity via over-expression of the ATP6V1C1 gene. Oral Oncol. 2008, 44, 193–199. [Google Scholar] [CrossRef]
- Sinning, A.; Liebmann, L.; Kougioumtzes, A.; Westermann, M.; Bruehl, C.; Hübner, C.A. Synaptic glutamate release is modulated by the Na+ -driven Cl-/HCO3− exchanger Slc4a8. J. Neurosci. 2011, 31, 7300–7311. [Google Scholar] [CrossRef]
- Herrero-Martín, M.D.; Pineda, M.; Briones, P.; López-Gallardo, E.; Carreras, M.; Benac, M.; Angel Idoate, M.; Vilaseca, M.A.; Artuch, R.; López-Pérez, M.J. A new pathologic mitochondrial DNA mutation in the cytochrome oxidase subunit I (MT-CO1). Hum. Mutat. 2008, 29, E112–E122. [Google Scholar] [CrossRef]
- Rouault, T.A.; Tang, C.K.; Kaptain, S.; Burgess, W.H.; Haile, D.J.; Samaniego, F.; Mcbride, O.W.; Harford, J.B.; Klausner, R.D. Cloning of the cDNA encoding an RNA regulatory protein--the human iron-responsive element-binding protein. Proc. Natl. Acad. Sci. USA 1990, 87, 7958–7962. [Google Scholar] [CrossRef] [Green Version]
- Curran, J.; Makara, M.A.; Little, S.; Musa, H.; Liu, B.; Wu, X.; Polina, I.; Alecusan, J.; Wright, P.J.; Li, J. EHD3-dependent endosome pathway regulates cardiac membrane excitability and physiology. Circ. Res. 2014, 115, 68–78. [Google Scholar] [CrossRef] [Green Version]
- Molloy, G.R.; Wilson, C.D.; Benfield, P.; De Vellis, J.; Kumar, S. Rat brain creatine kinase messenger RNA levels are high in primary cultures of brain astrocytes and oligodendrocytes and low in neurons. J. Neurochem. 1992, 59, 1925–1932. [Google Scholar] [CrossRef]
- Murthy, S.E.; Dubin, A.E.; Whitwam, T.; Jojoa-Cruz, S.; Cahalan, S.M.; Mousavi, S.A.R.; Ward, A.B.; Patapoutian, A. OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. Elife 2018, 7, e41844. [Google Scholar] [CrossRef]
- Sturchler, E.; Cox, J.A.; Durussel, I.; Weibel, M.; Heizmann, C.W. S100A16, a novel calcium-binding protein of the EF-hand superfamily. J. Biol. Chem. 2006, 281, 38905–38917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, W.; Qiao, L.; Petrenko, N.; Li, W.; Owens, A.T.; McDermott-Roe, C.; Musunuru, K. Functional annotation of TNNT2 variants of uncertain significance with genome-edited cardiomyocytes. Circulation 2018, 138, 2852–2854. [Google Scholar] [CrossRef] [PubMed]
- Reyes Fernandez, P.C.; Replogle, R.A.; Wang, L.; Zhang, M.; Fleet, J.C. Novel genetic loci control calcium absorption and femur bone mass as well as their response to low calcium intake in male bxd recombinant inbred mice. J. Bone Min. Res. 2016, 31, 994–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raffaello, A.; Mammucari, C.; Gherardi, G.; Rizzuto, R. Calcium at the center of cell signaling: Interplay between endoplasmic reticulum, mitochondria, and lysosomes. Trends Biochem. Sci. 2016, 41, 1035–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naro, F.; De Arcangelis, V.; Coletti, D.; Molinaro, M.; Zani, B.; Vassanelli, S.; Reggiani, C.; Teti, A.; Adamo, S. Increase in cytosolic Ca2+ induced by elevation of extracellular Ca2+ in skeletal myogenic cells. Am. J. Physiol. Cell Phsiol. 2003, 284, C969–C976. [Google Scholar] [CrossRef] [Green Version]







| Protein Name | Gene Name | FC 1 | p-Value |
|---|---|---|---|
| Upregulated | |||
| Tripartite motif containing 5 | TRIM5 | 4.60 | 0.001 |
| Aldehyde dehydrogenase 1 family member A1 | ALDH1A1 | 2.58 | 0.036 |
| Calcium/calmodulin dependent protein kinase II inhibitor 2 | CAMK2N2 | 2.06 | 0.005 |
| Myosin heavy chain 3 | MYH3 | 2.00 | 0.011 |
| Myosin light chain, phosphorylatable, fast skeletal muscle | MYLPF | 1.95 | 0.012 |
| ATP binding cassette subfamily B member 1 | ABCB1 | 1.90 | 0.035 |
| Mitochondrial amidoxime reducing component 2 | MARC2 | 1.75 | 0.036 |
| Troponin T2, cardiac type | TNNT2 | 1.74 | 0.012 |
| EH domain containing 3 | EHD3 | 1.67 | 0.004 |
| Cysteine and glycine-rich protein 3 | CSRP3 | 1.61 | 0.043 |
| Downregulated | |||
| Immunoglobulin superfamily containing leucine rich repeat | ISLR | 0.56 | 0.010 |
| Tyrosyl-trna synthetase 2 | YARS2 | 0.58 | 0.039 |
| Thymosin beta 15a | TMSB15A | 0.59 | 0.032 |
| Aldo-keto reductase family 1 member B | AKR1B1 | 0.59 | 0.025 |
| Fibrillin 2 | FBN2 | 0.61 | 0.007 |
| Tissue specific transplantation antigen P35B | TSTA3 | 0.68 | 0.000 |
| Armadillo repeat containing 9 | ARMC9 | 0.68 | 0.026 |
| Nuclear envelope integral membrane protein 1 | NEMP1 | 0.69 | 0.011 |
| Protein arginine methyltransferase 3 | PRMT3 | 0.69 | 0.024 |
| Collagen type XV alpha 1 chain | COL15A1 | 0.71 | 0.014 |
| Protein Name | Gene Name | FC 1 | p-Value |
|---|---|---|---|
| Glycerolipid metabolism | |||
| Aldehyde dehydrogenase 1 family member B1 | ALDH1B1 | 0.80 | 0.038 |
| Diacylglycerol kinase alpha | DGKA | 1.22 | 0.027 |
| Lysosome | |||
| Cathepsin C | CTSC | 1.37 | 0.040 |
| Galactosamine (N-acetyl)-6-sulfatase | GALNS | 1.37 | <0.001 |
| GM2 ganglioside activator | GM2A | 1.35 | 0.003 |
| GABAergic synapse | |||
| Gamma-aminobutyric acid receptor-associated protein-like 1 | LOC100518837 | 1.20 | 0.043 |
| Solute carrier family 38 member 5 | SLC38A5 | 0.83 | 0.033 |
| Phosphatidylinositol signaling system | |||
| Diacylglycerol kinase alpha | DGKA | 1.22 | 0.027 |
| Inositol polyphosphate-1-phosphatase | INPP1 | 1.25 | 0.031 |
| Valine, leucine and isoleucine degradation | |||
| Aldehyde dehydrogenase 1 family member B1 | ALDH1B1 | 0.80 | 0.038 |
| Methylcrotonoyl-coa carboxylase 2 | MCCC2 | 1.27 | <0.001 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, K.; Xu, D.; Wang, L.; Zhang, X.; Jiao, N.; Gong, L.; Yin, J. Association Analysis of Single-Cell RNA Sequencing and Proteomics Reveals a Vital Role of Ca2+ Signaling in the Determination of Skeletal Muscle Development Potential. Cells 2020, 9, 1045. https://doi.org/10.3390/cells9041045
Qiu K, Xu D, Wang L, Zhang X, Jiao N, Gong L, Yin J. Association Analysis of Single-Cell RNA Sequencing and Proteomics Reveals a Vital Role of Ca2+ Signaling in the Determination of Skeletal Muscle Development Potential. Cells. 2020; 9(4):1045. https://doi.org/10.3390/cells9041045
Chicago/Turabian StyleQiu, Kai, Doudou Xu, Liqi Wang, Xin Zhang, Ning Jiao, Lu Gong, and Jingdong Yin. 2020. "Association Analysis of Single-Cell RNA Sequencing and Proteomics Reveals a Vital Role of Ca2+ Signaling in the Determination of Skeletal Muscle Development Potential" Cells 9, no. 4: 1045. https://doi.org/10.3390/cells9041045
APA StyleQiu, K., Xu, D., Wang, L., Zhang, X., Jiao, N., Gong, L., & Yin, J. (2020). Association Analysis of Single-Cell RNA Sequencing and Proteomics Reveals a Vital Role of Ca2+ Signaling in the Determination of Skeletal Muscle Development Potential. Cells, 9(4), 1045. https://doi.org/10.3390/cells9041045