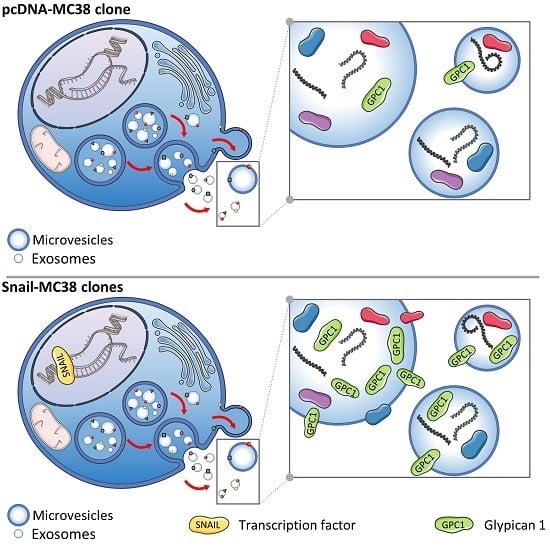
Glypican-1 Level Is Elevated in Extracellular Vesicles Released from MC38 Colon Adenocarcinoma Cells Overexpressing Snail
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. MC38 Nucleofection and Generation of Stable Clones
2.3. Confocal Microscopy
2.4. Proteolytic Activities of Matrix Metalloproteinases (MMPs)
2.5. Isolation of Extracellular Vesicles from Conditioned Media
2.6. Western Immunoblotting
2.7. RNA Isolation and Real-Time PCR Analysis
2.8. Transmission Electron Microscopy (TEM)
2.9. Size Distribution Analysis
2.10. Cell Proliferation Assay
2.11. Statistical Analysis
3. Results
3.1. Characterization of Snail-MC38 Stable Clones
3.2. Effect of Snail Overexpression in MC38 Cells on MMPs Activities and Gene Expression of Proteoglycans
3.3. Characterization of EVs Derived from MC38 Cells: PG Expression Profile
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CRC | Colorectal cancer |
| EMT | Epithelial-to-mesenchymal transition |
| EVs | Extracellular vesicles |
| GPC | Glypican |
| MMPs | Matrix Metalloproteinases |
| PGs | Proteoglycans |
| SDC | Syndecan |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Bueno, G.; Portillo, F.; Cano, A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene 2008, 27, 6958–6969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cano, C.; Motoo, Y.; Iovanna, J.L. Epithelial-to-Mesenchymal Transition in Pancreatic Adenocarcinoma. Sci. World J. 2010, 10, 1947–1957. [Google Scholar] [CrossRef]
- Santamaria, P.G.; Moreno-Bueno, G.; Portillo, F.; Cano, A. EMT: Present and future in clinical oncology. Mol. Oncol. 2017, 11, 718–738. [Google Scholar] [CrossRef] [Green Version]
- Nieto, M.A.; Huang, R.Y.Y.J.; Jackson, R.A.A.; Thiery, J.P.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef]
- Yang, J.; Weinberg, R.A. Epithelial-Mesenchymal Transition: At the Crossroads of Development and Tumor Metastasis. Dev. Cell 2008, 14, 818–829. [Google Scholar] [CrossRef] [Green Version]
- Przygodzka, P.; Papiewska-Pajak, I.; Bogusz, H.; Kryczka, J.; Sobierajska, K.; Kowalska, M.A.; Boncela, J. Neuromedin U is upregulated by Snail at early stages of EMT in HT29 colon cancer cells. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 2445–2453. [Google Scholar] [CrossRef]
- Przygodzka, P.; Papiewska-Pająk, I.; Bogusz-Koziarska, H.; Sochacka, E.; Boncela, J.; Kowalska, M.A. Regulation of miRNAs by Snail during epithelial-to-mesenchymal transition in HT29 colon cancer cells. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Kryczka, J.; Papiewska-Pajak, I.; Kowalska, M.A.; Boncela, J. Cathepsin B Is Upregulated and Mediates ECM Degradation in Colon Adenocarcinoma HT29 Cells Overexpressing Snail. Cells 2019, 8, 203. [Google Scholar] [CrossRef] [Green Version]
- Lane, R.E.; Korbie, D.; Hill, M.M.; Trau, M. Extracellular vesicles as circulating cancer biomarkers: Opportunities and challenges. Clin. Transl. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.A.; Purushothaman, A.; Ramani, V.C.; Vlodavsky, I.; Sanderson, R.D. Heparanase Regulates Secretion, Composition, and Function of Tumor Cell-derived Exosomes. J. Biol. Chem. 2013, 288, 10093–10099. [Google Scholar] [CrossRef] [Green Version]
- Purushothaman, A.; Bandari, S.K.; Liu, J.; Mobley, J.A.; Brown, E.A.; Sanderson, R.D. Fibronectin on the surface of myeloma cell-derived exosomes mediates exosome-cell interactions. J. Biol. Chem. 2016, 291, 1652–1663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roucourt, B.; Meeussen, S.; Bao, J.; Zimmermann, P.; David, G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015, 25, 412–428. [Google Scholar] [CrossRef] [Green Version]
- Sanderson, R.D.; Elkin, M.; Rapraeger, A.C.; Ilan, N.; Vlodavsky, I. Heparanase regulation of cancer, autophagy and inflammation: New mechanisms and targets for therapy. FEBS J. 2017, 284, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Théry, C. Cancer: Diagnosis by extracellular vesicles. Nature 2015, 523, 161–162. [Google Scholar] [CrossRef]
- Garnier, D.; Magnus, N.; Meehan, B.; Kislinger, T.; Rak, J. Qualitative changes in the proteome of extracellular vesicles accompanying cancer cell transition to mesenchymal state. Exp. Cell Res. 2013, 319, 2747–2757. [Google Scholar] [CrossRef] [Green Version]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef] [Green Version]
- Bland, C.L.; Byrne-Hoffman, C.N.; Fernandez, A.; Rellick, S.L.; Deng, W.; Klinke, D.J. Exosomes derived from B16F0 melanoma cells alter the transcriptome of cytotoxic T cells that impacts mitochondrial respiration. FEBS J. 2018, 285, 1033–1050. [Google Scholar] [CrossRef] [Green Version]
- Weidle, U.H.; Birzele, F.; Kollmorgen, G.; Rüger, R. The multiple roles of exosomes in metastasis. Cancer Genom. Proteom. 2017, 14, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nangami, G.; Koumangoye, R.; Shawn Goodwin, J.; Sakwe, A.M.; Marshall, D.; Higginbotham, J.; Ochieng, J. Fetuin-A associates with histones intracellularly and shuttles them to exosomes to promote focal adhesion assembly resulting in rapid adhesion and spreading in breast carcinoma cells. Exp. Cell Res. 2014, 328, 388–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Mathivanan, S.; Ji, H.; Simpson, R.J. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol. Cell. Proteom. 2013, 12, 587–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Chen, Y.; Guo, X.; Zhou, L.; Jia, Z.; Peng, Z.; Tang, Y.; Liu, W.; Zhu, B.; Wang, L.; et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J. Cell. Mol. Med. 2017. [Google Scholar] [CrossRef]
- Li, J.; Li, B.; Ren, C.; Chen, Y.; Guo, X.; Zhou, L.; Peng, Z.; Tang, Y.; Chen, Y.; Liu, W.; et al. The clinical significance of circulating GPC1 positive exosomes and its regulative miRNAs in colon cancer patients. Oncotarget 2017, 8, 101189–101202. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Karamanos, N.K. Proteoglycans remodeling in cancer: Underlying molecular mechanisms. Matrix Biol. 2019, 75–76, 220–259. [Google Scholar] [CrossRef]
- Filmus, J.; Capurro, M. Glypican-3: A marker and a therapeutic target in hepatocellular carcinoma. FEBS J. 2013, 280, 2471–2476. [Google Scholar] [CrossRef]
- Rapraeger, A.C. Synstatin: A selective inhibitor of the syndecan-1-coupled IGF1R-αvβ3 integrin complex in tumorigenesis and angiogenesis. FEBS J. 2013, 280, 2207–2215. [Google Scholar] [CrossRef]
- Rapraeger, A.C.; Ell, B.J.; Roy, M.; Li, X.; Morrison, O.R.; Thomas, G.M.; Beauvais, D.M. Vascular endothelial-cadherin stimulates syndecan-1-coupled insulin-like growth factor-1 receptor and cross-talk between αvβ3 integrin and vascular endothelial growth factor receptor 2 at the onset of endothelial cell dissemination during angiogenesis. FEBS J. 2013, 280, 2194–2206. [Google Scholar] [CrossRef] [Green Version]
- Karamanos, N.K.; Piperigkou, Z.; Theocharis, A.D.; Watanabe, H.; Franchi, M.; Baud, S.; Brézillon, S.; Götte, M.; Passi, A.; Vigetti, D.; et al. Proteoglycan chemical diversity drives multifunctional cell regulation and therapeutics. Chem. Rev. 2018, 118, 9152–9232. [Google Scholar] [CrossRef]
- Bandari, S.K.; Purushothaman, A.; Ramani, V.C.; Brinkley, G.J.; Chandrashekar, D.S.; Varambally, S.; Mobley, J.A.; Zhang, Y.; Brown, E.E.; Vlodavsky, I.; et al. Chemotherapy induces secretion of exosomes loaded with heparanase that degrades extracellular matrix and impacts tumor and host cell behavior. Matrix Biol. 2018, 65, 104–118. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stasiak, M.; Boncela, J.; Perreau, C.; Karamanou, K.; Chatron-Colliet, A.; Proult, I.; Przygodzka, P.; Chakravarti, S.; Maquart, F.X.; Kowalska, M.A.; et al. Lumican inhibits SNAIL-induced melanoma cell migration specifically by blocking MMP-14 activity. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef] [PubMed]
- Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, Ö.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; Bertier, L.; et al. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 2017, 14, 228–232. [Google Scholar] [CrossRef]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringnér, M.; Mörgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. USA 2013, 110, 7312–7317. [Google Scholar] [CrossRef] [Green Version]
- Vicente, C.M.; da Silva, D.A.; Sartorio, P.V.; Silva, T.D.; Saad, S.S.; Nader, H.B.; Forones, N.M.; Toma, L. Heparan Sulfate Proteoglycans in Human Colorectal Cancer. Anal. Cell. Pathol. 2018, 2018, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Yadav, L.; Puri, N.; Rastogi, V.; Satpute, P.; Ahmad, R.; Kaur, G. Matrix metalloproteinases and cancer - Roles in threat and therapy. Asian Pacific J. Cancer Prev. 2014, 15, 1085–1091. [Google Scholar] [CrossRef] [Green Version]
- Brézillon, S.; Pietraszek, K.; Maquart, F.X.; Wegrowski, Y. Lumican effects in the control of tumour progression and their links with metalloproteinases and integrins. FEBS J. 2013, 280, 2369–2381. [Google Scholar] [CrossRef]
- Pietraszek-Gremplewicz, K.; Karamanou, K.; Niang, A.; Dauchez, M.; Belloy, N.; Maquart, F.X.; Baud, S.; Brézillon, S. Small leucine-rich proteoglycans and matrix metalloproteinase-14: Key partners? Matrix Biol. 2019, 75–76, 271–285. [Google Scholar] [CrossRef]
- Pietraszek, K.; Chatron-Colliet, A.; Brézillon, S.; Perreau, C.; Jakubiak-Augustyn, A.; Krotkiewski, H.; Maquart, F.X.; Wegrowski, Y. Lumican: A new inhibitor of matrix metalloproteinase-14 activity. FEBS Lett. 2014, 588, 4319–4324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couchman, J.R.; Multhaupt, H.; Sanderson, R.D. Recent insights into cell surface heparan sulphate proteoglycans and cancer. F1000Research 2016, 5. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papiewska-Pająk, I.; Krzyżanowski, D.; Katela, M.; Rivet, R.; Michlewska, S.; Przygodzka, P.; Kowalska, M.A.; Brézillon, S. Glypican-1 Level Is Elevated in Extracellular Vesicles Released from MC38 Colon Adenocarcinoma Cells Overexpressing Snail. Cells 2020, 9, 1585. https://doi.org/10.3390/cells9071585
Papiewska-Pająk I, Krzyżanowski D, Katela M, Rivet R, Michlewska S, Przygodzka P, Kowalska MA, Brézillon S. Glypican-1 Level Is Elevated in Extracellular Vesicles Released from MC38 Colon Adenocarcinoma Cells Overexpressing Snail. Cells. 2020; 9(7):1585. https://doi.org/10.3390/cells9071585
Chicago/Turabian StylePapiewska-Pająk, Izabela, Damian Krzyżanowski, Maria Katela, Romain Rivet, Sylwia Michlewska, Patrycja Przygodzka, M. Anna Kowalska, and Stéphane Brézillon. 2020. "Glypican-1 Level Is Elevated in Extracellular Vesicles Released from MC38 Colon Adenocarcinoma Cells Overexpressing Snail" Cells 9, no. 7: 1585. https://doi.org/10.3390/cells9071585
APA StylePapiewska-Pająk, I., Krzyżanowski, D., Katela, M., Rivet, R., Michlewska, S., Przygodzka, P., Kowalska, M. A., & Brézillon, S. (2020). Glypican-1 Level Is Elevated in Extracellular Vesicles Released from MC38 Colon Adenocarcinoma Cells Overexpressing Snail. Cells, 9(7), 1585. https://doi.org/10.3390/cells9071585