Deep Characterization of Circular RNAs from Human Cardiovascular Cell Models and Cardiac Tissue
Abstract
:1. Introduction
2. Materials and Methods
2.1. hiPSCs Maintenance Culture
2.2. hiPSCs Differentiation towards Cardiac Myocytes (hiPSC-CMs)
2.3. HUVEC Cell Culture
2.4. Pig Tissue Samples
2.5. RNA Extraction from Human Hearts
2.6. RNA Extraction from HUVECs
2.7. RNA Extraction from hiPSC-CMs
2.8. RNA Sequencing
2.9. Study Approval
2.10. RNA-seq Read-Mapping
2.11. CircRNA Detection
2.12. Test for Enriched CircRNAs
2.13. Extraction of RNase R-Resistant CircRNA Exons
2.14. Identification of circRNAs Enriched for m6A-Methylation
3. Results
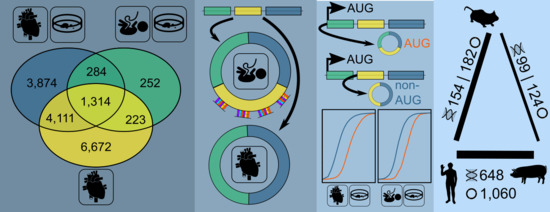
3.1. Expressed CircRNAs in Human Cardiovascular Cell Models and Cardiac Tissue
3.2. Internal CircRNA Structures
3.3. Potential Interaction of Subsets of CircRNAs
3.4. AUG CircRNAs in hiPSC-CMs and Cardiovascular Cell Models
3.5. CircRNA Conservation Throughout Multiple Species
4. Discussion
4.1. Assessment of CircRNA Interactions
4.2. AUG circRNAs Exhibit Specific Features
4.3. A Highly Conserved Subset of Cardiac CircRNAs
4.4. Deciphering CircRNA Sequences
4.5. Translational Aspects
4.6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| circRNA | circular RNA |
| HUVEC | Human umbilical vein endothelial cell |
| hiPSC-CM | Human induced pluripotent stem cell-derived cardiac myocyte |
| CVD | Cardiovascular disease |
| m6A | N6-methyladenosine |
| RBP | RNA binding protein |
| rRNA | Ribosomal RNA |
| FDR | False discovery rate |
| Ago2 | Argonaute 2 protein |
| BSJ | Back-splice junction |
| AUG | Methionine amino acid |
| CLIP | Cross-linking immunoprecipitation |
| UTR | Untranslated region |
| CDS | Coding sequence |
Appendix A





| Term | Adjusted p-Value |
|---|---|
| GO biological process enriched in HUVEC AUG circRNA host genes | |
| protein ubiquitination (GO:0016567) | 0.000008 |
| protein polyubiquitination (GO:0000209) | 0.000062 |
| protein modification by small protein conjugation (GO:0032446) | 0.000111 |
| ubiquitin-dependent protein catabolic process (GO:0006511) | 0.000707 |
| positive regulation of gene expression (GO:0010628) | 0.000808 |
| peptidyl-serine phosphorylation (GO:0018105) | 0.001548 |
| proteasomal protein catabolic process (GO:0010498) | 0.001578 |
| proteasome-mediated ubiquitin-dependent protein catabolic process (GO:0043161) | 0.006559 |
| positive regulation of transcription, DNA-templated (GO:0045893) | 0.007753 |
| phosphorylation (GO:0016310) | 0.009155 |
| GO biological process enriched in HUVEC non-AUG circRNA host genes | |
| protein modification by small protein conjugation (GO:0032446) | 0.000003 |
| protein ubiquitination (GO:0016567) | 0.000011 |
| regulation of translation (GO:0006417) | 0.000104 |
| protein polyubiquitination (GO:0000209) | 0.000133 |
| phosphorylation (GO:0016310) | 0.001204 |
| intracellular protein transport (GO:0006886) | 0.001390 |
| gene expression (GO:0010467) | 0.001411 |
| microtubule cytoskeleton organization involved in mitosis (GO:1902850) | 0.001602 |
| modification-dependent protein catabolic process (GO:0019941) | 0.001810 |
| regulation of coenzyme metabolic process (GO:0051196) | 0.001955 |
| GO biological process enriched in hiPSC-CM AUG circRNA host genes | |
| protein polyubiquitination (GO:0000209) | 0.005390 |
| protein ubiquitination (GO:0016567) | 0.010307 |
| peptidyl-serine phosphorylation (GO:0018105) | 0.046533 |
| cellular protein modification process (GO:0006464) | 0.053963 |
| GO biological process enriched in hiPSC-CM non-AUG circRNA host genes | |
| protein phosphorylation (GO:0006468) | 0.000000 |
| mitotic cell cycle phase transition (GO:0044772) | 0.000001 |
| phosphorylation (GO:0016310) | 0.000001 |
| cellular protein modification process (GO:0006464) | 0.000002 |
| cellular response to DNA damage stimulus (GO:0006974) | 0.000002 |
| protein modification by small protein conjugation (GO:0032446) | 0.000012 |
| DNA metabolic process (GO:0006259) | 0.000024 |
| regulation of translation (GO:0006417) | 0.000025 |
| DNA repair (GO:0006281) | 0.000049 |
| RNA metabolic process (GO:0016070) | 0.000068 |
| Term | Adjusted p-Value |
|---|---|
| GO molecular function enriched in HUVEC AUG circRNA host genes | |
| ubiquitin-protein transferase activity (GO:0004842) | 0.000004 |
| ubiquitin-like protein ligase activity (GO:0061659) | 0.004251 |
| cadherin binding (GO:0045296) | 0.008171 |
| protein serine/threonine kinase activity (GO:0004674) | 0.010463 |
| ubiquitin-protein ligase activity (GO:0061630) | 0.010496 |
| SUMO binding (GO:0032183) | 0.014643 |
| N6-methyladenosine-containing RNA binding (GO:1990247) | 0.046624 |
| kinase binding (GO:0019900) | 0.056694 |
| RNA binding (GO:0003723) | 0.058371 |
| protein kinase activity (GO:0004672) | 0.061175 |
| GO molecular function enriched in HUVEC non-AUG circRNA host genes | |
| RNA binding (GO:0003723) | 0.000000 |
| ubiquitin-like protein ligase activity (GO:0061659) | 0.000000 |
| ubiquitin-protein ligase activity (GO:0061630) | 0.000000 |
| ubiquitin-protein transferase activity (GO:0004842) | 0.000001 |
| Ras GTPase binding (GO:0017016) | 0.000032 |
| kinase activity (GO:0016301) | 0.000048 |
| cadherin binding (GO:0045296) | 0.000065 |
| protein kinase activity (GO:0004672) | 0.000098 |
| aminoacyl-tRNA ligase activity (GO:0004812) | 0.000131 |
| protein serine/threonine kinase activity (GO:0004674) | 0.000285 |
| GO molecular function enriched in hiPSC-CM AUG circRNA host genes | |
| ubiquitin-protein transferase activity (GO:0004842) | 0.017289 |
| protein serine/threonine kinase activity (GO:0004674) | 0.047033 |
| lipase activity (GO:0016298) | 0.082577 |
| RNA polymerase II transcription factor binding (GO:0001085) | 0.088788 |
| phosphatase binding (GO:0019902) | 0.094872 |
| activating transcription factor binding (GO:0033613) | 0.096428 |
| RNA polymerase II activating transcription factor binding (GO:0001102) | 0.096574 |
| GO molecular function enriched in hiPSC-CM non-AUG circRNA host genes | |
| protein serine/threonine kinase activity (GO:0004674) | 0.000000 |
| RNA binding (GO:0003723) | 0.000000 |
| protein kinase activity (GO:0004672) | 0.000001 |
| ubiquitin-protein ligase activity (GO:0061630) | 0.000017 |
| aminoacyl-tRNA ligase activity (GO:0004812) | 0.000062 |
| ubiquitin-like protein ligase activity (GO:0061659) | 0.000069 |
| kinase activity (GO:0016301) | 0.000073 |
| ATPase activity (GO:0016887) | 0.000078 |
| adenyl ribonucleotide binding (GO:0032559) | 0.000215 |
| ubiquitin-protein transferase activity (GO:0004842) | 0.000219 |
References
- Abubakar, I.I.; Tillmann, T.; Banerjee, A. GBD 2013 Mortality and Causes of Death Collaborators Global, regional, and national age–sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015, 385, 117–171. [Google Scholar] [CrossRef]
- Maron, B.J.; Maron, M.S.; Semsarian, C. Genetics of hypertrophic cardiomyopathy after 20 years: Clinical perspectives. J. Am. Coll. Cardiol. 2012, 60, 705–715. [Google Scholar] [CrossRef] [Green Version]
- McPherson, R.; Tybjaerg-Hansen, A. Genetics of Coronary Artery Disease. Circ. Res. 2016, 118, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.S.; Cooper, T.A. Splicing in disease: Disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007, 8, 749–761. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Lasda, E.; Parker, R.O.Y. Circular RNAs: Diversity of form and function. RNA 2014, 1829–1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Siede, D.; Rapti, K.; Gorska, A.A.; Katus, H.A.; Altmüller, J.; Boeckel, J.N.; Meder, B.; Maack, C.; Völkers, M.; Müller, O.J.; et al. Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J. Mol. Cell. Cardiol. 2017, 109, 48–56. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [Green Version]
- Devaux, Y.; Creemers, E.E.; Boon, R.A.; Werfel, S.; Thum, T.; Engelhardt, S.; Dimmeler, S.; Squire, I.; Network, C. Circular RNAs in heart failure. Eur. J. Heart Fail. 2017, 19, 701–709. [Google Scholar] [CrossRef] [Green Version]
- Memczak, S.; Papavasileiou, P.; Peters, O.; Rajewsky, N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE 2015, 10, e0141214. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Zang, J.; Lu, D.; Xu, A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J. Neurosci. Res. 2018, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef] [Green Version]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakobi, T.; Dieterich, C. Computational approaches for circular RNA analysis. Wiley Interdiscip. Rev. RNA 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Hansen, T.B. Improved circRNA Identification by Combining Prediction Algorithms. Front. Cell Dev. Biol. 2018, 6, 1–9. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Jakobi, T.; Czaja-Hasse, L.F.; Reinhardt, R.; Dieterich, C. Profiling and Validation of the Circular RNA Repertoire in Adult Murine Hearts. Genom. Proteom. Bioinform. 2016, 14, 216–223. [Google Scholar] [CrossRef] [Green Version]
- Werfel, S.; Nothjunge, S.; Schwarzmayr, T.; Strom, T.M.M.; Meitinger, T.; Engelhardt, S. Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell. Cardiol. 2016, 98, 103–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.A.; Reckman, Y.J.; Aufiero, S.; Van Den Hoogenhof, M.M.; Van Der Made, I.; Beqqali, A.; Koolbergen, D.R.; Rasmussen, T.B.; Van Der Velden, J.; Creemers, E.E.; et al. RBM20 Regulates Circular RNA Production from the Titin Gene. Circ. Res. 2016, 119, 996–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, W.L.; Lim, B.T.; Anene-Nzelu, C.G.; Ackers-Johnson, M.; Dashi, A.; See, K.; Tiang, Z.; Lee, D.P.; Chua, W.W.; Luu, T.D.; et al. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017, 113, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Versari, D.; Daghini, E.; Virdis, A.; Ghiadoni, L.; Taddei, S. Endothelial dysfunction as a target for prevention of cardiovascular disease. Diabetes Care 2009, 32, S314–S321. [Google Scholar] [CrossRef] [Green Version]
- Medina-Leyte, D.J.; Domínguez-Pérez, M.; Mercado, I.; Villarreal-Molina, M.T.; Jacobo-Albavera, L. Use of human umbilical vein endothelial cells (HUVEC) as a model to study cardiovascular disease: A review. Appl. Sci. 2020, 10, 938. [Google Scholar] [CrossRef] [Green Version]
- Van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.L.; et al. The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260.e29. [Google Scholar] [CrossRef] [Green Version]
- Kmietczyk, V.; Riechert, E.; Kalinski, L.; Boileau, E.; Malovrh, E.; Malone, B.; Gorska, A.; Hofmann, C.; Varma, E.; Jürgensen, L.; et al. M 6 A-mRNA methylation regulates cardiac gene expression and cellular growth. Life Sci. Alliance 2019, 2, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Spengler, R.M.; Zhang, X.; Cheng, C.; McLendon, J.M.; Skeie, J.M.; Johnson, F.L.; Davidson, B.L.; Boudreau, R.L. Elucidation of transcriptome-wide microRNA binding sites in human cardiac tissues by Ago2 HITS-CLIP. Nucleic Acids Res. 2016, 44, 7120–7131. [Google Scholar] [CrossRef]
- Lian, X.; Hsiao, C.; Wilson, G.; Zhu, K.; Hazeltine, L.B.; Azarin, S.M.; Raval, K.K.; Zhang, J.; Kamp, T.J.; Palecek, S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 2012, 109. [Google Scholar] [CrossRef] [Green Version]
- Lian, X.; Zhang, J.; Azarin, S.M.; Zhu, K.; Hazeltine, L.B.; Bao, X.; Hsiao, C.; Kamp, T.J.; Palecek, S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 2013, 8, 162–175. [Google Scholar] [CrossRef] [Green Version]
- Pleger, S.T.; Shan, C.; Ksienzyk, J.; Bekeredjian, R.; Boekstegers, P.; Hinkel, R.; Schinkel, S.; Leuchs, B.; Ludwig, J.; Qiu, G.; et al. Cardiac AAV9-S100A1 gene therapy rescues post-ischemic heart failure in a preclinical large animal model. Sci. Transl. Med. 2011, 3, 92ra64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roehr, J.T.; Dieterich, C.; Reinert, K. Flexbar 3.0–SIMD and multicore parallelization. Bioinformatics 2017, 33, 2941–2942. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Metge, F.; Dieterich, C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics 2015. [Google Scholar] [CrossRef] [Green Version]
- Jakobi, T.; Uvarovskii, A.; Dieterich, C. circtools—A one-stop software solution for circular RNA research. Bioinformatics 2019, 35, 2326–2328. [Google Scholar] [CrossRef] [Green Version]
- Jakobi, T.; Dieterich, C. Deep computational circular RNA analytics from RNA-seq data. In Circular RNAs; Springer: Berlin/Heidelberg, Germany, 2018; pp. 9–25. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Wang, L.; Ren, X.; Zhang, Y.; Zhang, H. LRPPRC: A multifunctional protein involved in energy metabolism and human disease. Front. Physiol. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Hong, H.Q.; Lu, J.; Fang, X.L.; Zhang, Y.H.; Cai, Y.; Yuan, J.; Liu, P.Q.; Ye, J.T. G3BP2 is involved in isoproterenol-induced cardiac hypertrophy through activating the NF-κB signaling pathway. Acta Pharmacol. Sin. 2018, 39, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Burnatowska-Hledin, M.; Lazdins, I.B.; Listenberger, L.; Zhao, P.; Sharangpani, A.; Folta, V.; Card, B. VACM-1 receptor is specifically expressed in rabbit vascular endothelium and renal collecting tubule. Am. J. Physiol. Ren. Physiol. 1999, 276, 199–209. [Google Scholar] [CrossRef]
- Gupta, S.K.; Garg, A.; Bär, C.; Chatterjee, S.; Foinquinos, A.; Milting, H.; Streckfus-Bomeke, K.; Fiedler, J.; Thum, T. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression short communication. Circ. Res. 2018, 122, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Kazim, N.; Adhikari, A.; Davie, J. The transcription elongation factor TCEA3 promotes the activity of the myogenic regulatory factors. PLoS ONE 2019, 14, e0217680. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Kendal, J.K.; MacIsaac, R.M.; Demetrick, D.J. WSB1: From homeostasis to hypoxia. J. Biomed. Sci. 2016, 23, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N 6 -methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, T.B.; Aliwarga, E.; Luu, T.D.A.; Li, Y.P.; Ng, S.L.; Annadoray, L.; Sian, S.; Ackers-Johnson, M.A.; Foo, R.S.Y. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc. Res. 2019, 1998–2007. [Google Scholar] [CrossRef] [Green Version]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, 451–454. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, 1–38. [Google Scholar] [CrossRef]
- Stagsted, L.V.; Nielsen, K.M.; Daugaard, I.; Hansen, T.B. Noncoding AUG circRNAs constitute an abundant and conserved subclass of circles. Life Sci. Alliance 2019, 2, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Chao, C.W.; Chan, D.C.; Kuo, A.; Leder, P. The mouse formin (Fmn) gene: Abundant circular RNA transcripts and gene- targeted deletion analysis. Mol. Med. 1998, 4, 614–628. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Vliegen, H.; Van der Laarse, A.; Cornelisse, C.; Eulderink, F. Myocardial changes in pressure overload-induced left ventricular hypertrophy: A study on tissue composition, polyploidization and multinucleation. Eur. Heart J. 1991, 12, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.X.; Chen, X.; Xia, L.P.; Zhang, J.X.; Pan, Z.Z.; Ma, X.D.; Han, K.; Chen, J.W.; Judde, J.G.; Deas, O.; et al. N 6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B.; et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cell 2019, 76, 96–109.e9. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Long, B.; Liu, F.; Wang, J.X.; Liu, C.Y.; Zhao, B.; Zhou, L.Y.; Sun, T.; Wang, M.; Yu, T.; et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016, 37, 2602a–2611a. [Google Scholar] [CrossRef]
- Vausort, M.; Salgado-Somoza, A.; Zhang, L.; Leszek, P.; Scholz, M.; Teren, A.; Burkhardt, R.; Thiery, J.; Wagner, D.R.; Devaux, Y. Myocardial Infarction-Associated Circular RNA Predicting Left Ventricular Dysfunction. J. Am. Coll. Cardiol. 2016, 68, 1247–1248. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, Y.; Xu, S.; Liu, Y.; Yu, L.; Li, Z.; Xue, X.; Wang, H. Plasma Circular RNAs, Hsa_circRNA_025016, Predict Postoperative Atrial Fibrillation After Isolated Off-Pump Coronary Artery Bypass Grafting. J. Am. Heart Assoc. 2018, 7, e006642. [Google Scholar] [CrossRef] [Green Version]
- Vilades, D.; Martínez-Camblor, P.; Ferrero-Gregori, A.; Bär, C.; Lu, D.; Xiao, K.; Vea, A.; Nasarre, L.; Vega, J.S.; Leta, R.; et al. Plasma circular RNA hsa_circ_0001445 and coronary artery disease: Performance as a biomarker. Faseb J. 2020, 34, 4403–4414. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Xu, J.D.; Fang, X.H.; Zhu, J.N.; Yang, J.; Pan, R.; Yuan, S.J.; Zeng, N.; Yang, Z.Z.; Yang, H.; et al. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc. Res. 2020, 116, 1323–1334. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Chen, Y.; Wu, Z.K.; Foster, F.S.; Yang, Z.; Li, X.; Yang, B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017, 38, 1402–1412. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Du, W.W.; Wu, Y.; Yang, Z.; Awan, F.M.; Li, X.; Yang, W.; Zhang, C.; Yang, Q.; Yee, A.; et al. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics 2017, 7, 3842–3855. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.H.; Li, R.; Su, Y.M.; Xiao, J.; Pan, M.; Cai, X.X.; Ji, X.P. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS ONE 2016, 11, e0151753. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Qi, B.; Wu, X.; Peng, S.; Zhou, G.; Wei, Y.; Xu, J.; Chen, S.; Liu, S. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J. Mol. Cell. Cardiol. 2019, 130, 10–22. [Google Scholar] [CrossRef]
- Huang, S.; Li, X.; Zheng, H.; Si, X.; Li, B.; Wei, G.; Li, C.; Chen, Y.; Chen, Y.; Liao, W.; et al. Loss of Super-Enhancer-Regulated circRNA Nfix Induces Cardiac Regeneration After Myocardial Infarction in Adult Mice. Circulation 2019, 139, 2857–2876. [Google Scholar] [CrossRef] [PubMed]






| Gene | Gene Description | Note |
|---|---|---|
| PRRC2C | Proline Rich Coiled-Coil 2C | - |
| EPS15 * | Epidermal Growth Factor Receptor Pathway Substrate 15 | Highly expressed in heart |
| TBC1D8 | TBC1 Domain Family Member 8 | - |
| NDUFS1 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | Highest expression in heart |
| LRPPRC | Leucine Rich Pentatricopeptide Repeat Containing | Multifunctional Protein Involved in Energy Metabolism and Human Disease [39] |
| ESYT2 * | Extended Synaptotagmin 2 | Tethers the endoplasmic reticulum to the cell membrane and promotes the formation of appositions between the endoplasmic reticulum and the cell membrane |
| SEC62 * | SEC62 Homolog, Preprotein Translocation Factor | Mediates post-translational transport of precursor polypeptides across endoplasmic reticulum |
| NOP14-AS1 | NOP14 Antisense RNA 1 | LncRNA |
| G3BP2 | G3BP Stress Granule Assembly Factor 2 | Involved in isoprenaline-induced cardiac hypertrophy [40] |
| CUL5 * | Cullin 5 | Protein is expressed at its highest levels in heart and skeletal tissue [41] |
| STRN3 | Striatin 3 | Regulation through Quaking [42] |
| USP7 | Ubiquitin Specific Peptidase 7 | - |
| TCEA1 | Transcription Elongation Factor A1 | Promotes activity of the myogenic regulatory factors in skeletal muscle [43] |
| ATP5F1C * | ATP Synthase F1 Subunit Gamma | Highly expressed in heart |
| RNF10 | Ring Finger Protein 10 | Highly expressed in heart |
| WSB1 | WD Repeat and SOCS Box Containing 1 | Linked to hypoxia response [44] |
| SPAG9 | Sperm Associated Antigen 9 | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jakobi, T.; Siede, D.; Eschenbach, J.; Heumüller, A.W.; Busch, M.; Nietsch, R.; Meder, B.; Most, P.; Dimmeler, S.; Backs, J.; et al. Deep Characterization of Circular RNAs from Human Cardiovascular Cell Models and Cardiac Tissue. Cells 2020, 9, 1616. https://doi.org/10.3390/cells9071616
Jakobi T, Siede D, Eschenbach J, Heumüller AW, Busch M, Nietsch R, Meder B, Most P, Dimmeler S, Backs J, et al. Deep Characterization of Circular RNAs from Human Cardiovascular Cell Models and Cardiac Tissue. Cells. 2020; 9(7):1616. https://doi.org/10.3390/cells9071616
Chicago/Turabian StyleJakobi, Tobias, Dominik Siede, Jessica Eschenbach, Andreas W. Heumüller, Martin Busch, Rouven Nietsch, Benjamin Meder, Patrick Most, Stefanie Dimmeler, Johannes Backs, and et al. 2020. "Deep Characterization of Circular RNAs from Human Cardiovascular Cell Models and Cardiac Tissue" Cells 9, no. 7: 1616. https://doi.org/10.3390/cells9071616
APA StyleJakobi, T., Siede, D., Eschenbach, J., Heumüller, A. W., Busch, M., Nietsch, R., Meder, B., Most, P., Dimmeler, S., Backs, J., Katus, H. A., & Dieterich, C. (2020). Deep Characterization of Circular RNAs from Human Cardiovascular Cell Models and Cardiac Tissue. Cells, 9(7), 1616. https://doi.org/10.3390/cells9071616