Molecular Protein and Expression Profile in the Primary Tumors of Clear Cell Renal Carcinoma and Metastases
Abstract
:1. Introduction
2. Materials and Methods
2.1. RNA Extraction
2.2. Determination of Expression Levels of AKT/M-TOR Signaling Pathway Components
2.3. Statistical Analysis
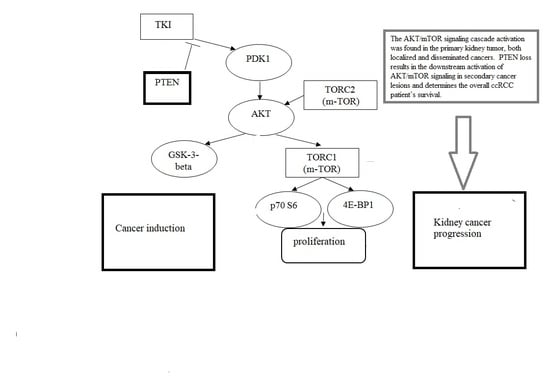
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Seyfried, T.N.; Huysentruyt, L.C. On the origin of cancer metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubo, H.; Mensurado, S.; Gonçalves-Sousa, N.; Serre, K.; Silva-Santos, B. Primary Tumors Limit Metastasis Formation through Induction of IL15-Mediated Cross-Talk between Patrolling Monocytes and NK Cells. Cancer Immunol. Res. 2017, 5, 812–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanin, L.; Rose, J. Suppression of Metastasis by Primary Tumor and Acceleration of Metastasis Following Primary Tumor Resection: A Natural Law? Bull. Math. Boil. 2018, 80, 519–539. [Google Scholar] [CrossRef] [PubMed]
- Vignot, S.; Besse, B.; Andre, F.; Spano, J.-P.; Soria, J.-C. Discrepancies between primary tumor and metastasis: A literature review on clinically established biomarkers. Crit. Rev. Oncol. 2012, 84, 301–313. [Google Scholar] [CrossRef]
- Szekely, B.; Nagy, Z.I.; Farago, Z.; Kiss, O.; Lotz, G.; Kovacs, K.A.; Madaras, L.; Udvarhelyi, N.; Dank, M.; Szentmartoni, G.; et al. Comparison of immunophenotypes of primary breast carcinomas and multiple corresponding distant metastases: An autopsy study of 25 patients. Clin. Exp. Metastasis 2016, 34, 103–113. [Google Scholar] [CrossRef]
- Dagher, J.; Kammerer-Jacquet, S.-F.; Dugay, F.; Beaumont, M.; Lespagnol, A.; Cornevin, L.; Verhoest, G.; Bensalah, K.; Rioux-Leclercq, N.; Belaud-Rotureau, M.-A. Clear cell renal cell carcinoma: A comparative study of histological and chromosomal characteristics between primary tumors and their corresponding metastases. Virchows Archiv 2017, 471, 107–115. [Google Scholar] [CrossRef]
- De Velasco, G.; Wankowicz, S.A.; Madison, R.; Ali, S.M.; Norton, C.; Duquette, A.; Ross, J.S.; Bossé, D.; Lalani, A.-K.A.; Miller, V.A.; et al. Targeted genomic landscape of metastases compared to primary tumours in clear cell metastatic renal cell carcinoma. Br. J. Cancer 2018, 118, 1238–1242. [Google Scholar] [CrossRef] [Green Version]
- Miikkulainen, P.; Högel, H.; Rantanen, K.; Suomi, T.; Kouvonen, P.; Elo, L.L.; Jaakkola, P. HIF prolyl hydroxylase PHD3 regulates translational machinery and glucose metabolism in clear cell renal cell carcinoma. Cancer Metab. 2017, 5, 5. [Google Scholar] [CrossRef] [Green Version]
- Mehdi, A.; Riazalhosseini, Y. Epigenome Aberrations: Emerging Driving Factors of the Clear Cell Renal Cell Carcinoma. Int. J. Mol. Sci. 2017, 18, 1774. [Google Scholar] [CrossRef] [Green Version]
- Pasha, M.; Siveen, K.; Frantz, R.; Agouni, A.; Munusamy, S. Metformin Induces Different Responses in Clear Cell Renal Cell Carcinoma Caki Cell Lines. Biomolecules 2019, 9, 113. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Yang, Z.; Xu, Y.-Q.; Chen, Y.; Yu, Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol. Cancer 2015, 14, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Que, W.-C.; Qiu, H.-Q.; Cheng, Y.; Liu, M.-B.; Wu, C.-Y.; Chen, Y. PTEN in kidney cancer: A review and meta-analysis. Clin. Chim. Acta 2018, 480, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Heinzelmann, J.; Unrein, A.; Wickmann, U.; Baumgart, S.; Stapf, M.; Szendrői, A.; Grimm, M.-O.; Gajda, M.R.; Wunderlich, H.; Junker, K. MicroRNAs with Prognostic Potential for Metastasis in Clear Cell Renal Cell Carcinoma: A Comparison of Primary Tumors and Distant Metastases. Ann. Surg. Oncol. 2013, 21, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Hager, M.; Haufe, H.; Alinger, B.; Kolbitsch, C. pS6 Expression in Normal Renal Parenchyma, Primary Renal Cell Carcinomas and their Metastases. Pathol. Oncol. Res. 2011, 18, 277–283. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Grönroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Cancer Genome Atlas Research Network. Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef] [Green Version]
- Semeniuk-Wojtaś, A.; Stec, R.; Szczylik, C. Are primary renal cell carcinoma and metastases of renal cell carcinoma the same cancer? Urol. Oncol. Semin. Orig. Investig. 2016, 34, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Biswas, S.; Troy, H.; Leek, R.; Chung, Y.L.; Li, J.L.; Raval, R.R.; Turley, H.; Gatter, K.; Pezzella, F.; Griffiths, J.R.; et al. Effects of HIF-1alpha and HIF2alpha on Growth and Metabolism of Clear-Cell Renal Cell Carcinoma 786-0 Xenografts. J. Oncol. 2010, 2010, 757908. [Google Scholar] [CrossRef]
- Wang, Q.; Qin, J.; Liu, X. Analysis of efficacy of sorafenib combined with vascular endothelial growth factor inhibitor on renal cell carcinoma. J. BUON 2019, 24, 1638–1643. [Google Scholar]
- Kitano, H.; Kitadai, Y.; Teishima, J.; Yuge, R.; Shinmei, S.; Goto, K.; Inoue, S.; Hayashi, T.; Sentani, K.; Yasui, W.; et al. Combination therapy using molecular-targeted drugs modulates tumor microenvironment and impairs tumor growth in renal cell carcinoma. Cancer Med. 2017, 6, 2308–2320. [Google Scholar] [CrossRef] [Green Version]
- Spirina, L.V.; Kondakova, I.V.; Tarasenko, N.V.; Slonimskaya, E.M.; A Usynin, E.; Gorbunov, A.K.; A Yurmazov, Z.; Chigevskaya, S.Y. Targeting of the AKT/m-TOR Pathway: Biomarkers of Resistance to Cancer Therapy—AKT/m-TOR Pathway and Resistance to Cancer Therapy. Zhongguo Fei Ai Za Zhi 2018, 21, 63–66. [Google Scholar] [PubMed]
- Spirina, L.V.; A Usynin, E.; A Yurmazov, Z.; Slonimskaya, E.M.; Kondakova, I.V. Effect of Targeted Therapy with Pazopanib on Expression Levels of Transcription, Growth Factors and Components of AKT/m-TOR Signaling Pathway in Patients with Renal Cell Carcinoma. Asian Pac. J. Cancer Prev. 2017, 18, 2977–2983. [Google Scholar] [PubMed]
- Poletto, V.; Rosti, V.; Biggiogera, M.; Guerra, G.; Moccia, F.; Porta, C. The role of endothelial colony forming cells in kidney cancer’s pathogenesis, and in resistance to anti-VEGFR agents and mTOR inhibitors: A speculative review. Crit. Rev. Oncol. 2018, 132, 89–99. [Google Scholar] [CrossRef]
- Spirina, L.V.; Usynin, Y.A.; Yurmazov, Z.A.; Slonimskaya, E.M.; Kolegova, E.S.; Kondakova, I.V. Transcription factors NF-kB, HIF-1, HIF-2, growth factor VEGF, VEGFR2 and carboanhydrase IX mRNA and protein level in the development of kidney cancer metastasis. Mol. Boil. 2017, 51, 328–332. [Google Scholar] [CrossRef]


| Gene | Amplicon | Sequence |
|---|---|---|
| CAIX NM_001216.2 | 217 bp | F 5′-GTTGCTGTCTCGCTTGGAA-3′ R 5′-CAGGGTGTCAGAGAGGGTGT-3′ |
| HIF-1α NM_001243084.1 | 188 bp | F 5′- CAAGAACCTACTGCTAATGCCA-3′ R 5′- TTTGGTGAGGCTGTCCGA-3′ |
| EPAS1 NM_001430.4 | 265 bp | F 5′- TGGAGTATGAAGAGCAAGCCT-3′ R 5′-GGGAACCTGCTCTTGCTGT-3′ |
| NFKB1 NM_001165412.1 | 144 bp | F 5′-CGTGTAAACCAAAGCCCTAAA-3′ R 5′-AACCAAGAAAGGAAGCCAAGT-3′ |
| RELA NM_001145138.1 | 271 bp | F 5′-GGAGCACAGATACCACCAAGA-3′ R 5′-GGGTTGTTGTTGGTCTGGAT-3′ |
| PTEN NM_001304717.2 | 136 bp | F 5′-GGGAATGGAGGGAATGCT-3′ R 5′-CGCAAACAACAAGCAGTGA-3′ |
| VEGFA NM_001025366.2 | 316 bp | F 5′-AGGGCAGAATCATCACGAA-3′ R 5′-TCTTGCTCTATCTTTCTTTGGTCT-3′ |
| KDR NM_002253.2 | 306 bp | F 5′-AACACAGCAGGAATCAGTCA-3′ R 5′-GTGGTGTCTGTGTCATCGGA-3′ |
| 4EBP1 NM_004095.3 | 244 bp | F 5′- CAGCCCTTTCTCCCTCACT -3′ R 5′- TTCCCAAGCACATCAACCT -3′ |
| AKT1 NM_001014431.1 | 181 bp | F 5′- CGAGGACGCCAAGGAGA-3′ R 5′- GTCATCTTGGTCAGGTGGTGT-3′ |
| C-RAF NM_002880.3 | 152 bp | F 5′- TGGTGTGTCCTGCTCCCT-3′ R 5′- ACTGCCTGCTACCTTACTTCCT-3′ |
| GSK3b NM_001146156.1 | 267 bp | F 5′- AGACAAGGACGGCAGCAA-3′ R 5′- CTGGAGTAGAAGAAATAACGCAAT-3′ |
| 70S kinase alpha NM_001272042.1 | 244 bp | F 5′- CAGCACAGCAAATCCTCAGA-3′ R 5′- ACACATCTCCCTCTCCACCTT-3′ |
| m-TOR NM_004958.3 | 160 bp | F 5′- CCAAAGGCAACAAGCGAT-3′ R 5′- TTCACCAAACCGTCTCCAA-3′ |
| PDK1 NM_001278549.1 | 187 bp | F 5′- TCACCAGGACAGCCAATACA-3′ R 5′- CTCCTCGGTCACTCATCTTCA-3′ |
| GAPDH NM_001256799.2 | 138 bp | F 5′-GGAAGTCAGGTGGAGCGA-3′ R 5′-GCAACAATATCCACTTTACCAGA-3′ |
| Indicator | Normal Renal Parenchyma | Localized Kidney Cancer Tissue | Disseminated Kidney Cancer Tissue | RCC Metastatic Tissue |
|---|---|---|---|---|
| Content of NF-κB p50, NF-κB p65, HIF-1, and HIF-2 | ||||
| NF-κB p50, RLU/mg of protein | 4.16 (3.76–4.66) | 7.6 (5.5–15.0) * | 6.7 (4.8–25.4) * | 6.55 (4.7–12.4) * |
| Kruskal–Wallis Test: p = 0.0234 | ||||
| NF-κB p65, RLU/mg of protein | 4.88 (4.66–5.21) | 7.5 (4.0–13.0) * | 6.9 (6.3–21.5) * | 3.3 (2.45–5.2) **, *** |
| Kruskal–Wallis Test: p = 0.0019 | ||||
| HIF-1α, RlLU/mg of protein | 0.98 (0.62–1.18) | 4.7 (2.2–7.8) * | 5.4 (2.9–11.2) * | 1.75 (1.55–2.45) *, **, *** |
| Kruskal–Wallis Test: p = 0.0001 | ||||
| HIF-2, pg/mg of protein | 248.9 (155.9–464.0) | 364.5 (242.7–1308.0) * | 334.9 (296.8–396.5) | 1250.1 (860.8–1750.4) *, **, *** |
| Kruskal–Wallis Test: p = 0.0984 | ||||
| mRNA level of NF-κB p50, NF-κB p65, HIF-1, and HIF-2 | ||||
| NF-κB p50, Relative Units | Used In Calculation | 0.13 (0.08; 6.36) | 2.00 (0.06; 13.55) | 6.00 (2.05; 28.14) |
| Kruskal–Wallis Test: p = 0.3289 | ||||
| NF-κB p65, Relative Units | Used In Calculation | 1.29 (0.08; 2.00) | 1.00 (0.26; 8.00) | 4.11 (0.14; 16.24) |
| Kruskal–Wallis Test: p = 0.6452 | ||||
| HIF-1, Relative Units | Used In Calculation | 1.60 (0.50; 4.00) | 0.50 (0.50; 1.80) | 20.80 (9.60; 35.30) **,*** |
| Kruskal–Wallis Test: p = 0.0202 | ||||
| HIF-2, Relative Units | Used In Calculation | 1.0 (0.50; 4.00) | 1.00 (0.06; 6.60) | 2.95 (1.24; 12.27) |
| Kruskal–Wallis Test: p = 0.8958 | ||||
| Indicator | Normal Renal Parenchyma | Localized Kidney Cancer Tissue | Disseminated Kidney Cancer Tissue | RCC Metastatic Tissue |
|---|---|---|---|---|
| Content of mRNA level of VEGF, VEGFR2, and CAIX | ||||
| VEGF, pg/mg of protein | 10.28 (8.46; 14.3) | 54.1 (19.7; 87.9) * | 68.9 (11.1; 220.0) * | 58.8 (44.0; 87.8) * *** |
| Kruskal–Wallis test: p = 0.0001 | ||||
| VEGFR2, pg/mg of protein | 39.64 (22.3; 48.8) | 35.0 (20.0; 53.0) | 62.5 (28.2; 74.3) *, ** | 90.5 (35.6; 110.5) *, *** |
| Kruskal–Wallis test: p = 1.000 | ||||
| CAIX, pg/mg of protein | 246.9 (111.1; 523.6) | 292.2 (223.8; 1525.4) | 189.3 (100.0; 199.2) ** | 62.4 (30.5; 78.8) *, **, *** |
| Kruskal–Wallis test: p = 0.0349 | ||||
| mRNA level of VEGF, VEGFR2, and CAIX | ||||
| VEGF, Relative Units | used in calculation | 0.63 (0.02; 15.80) | 1.03 (0.13; 42.20) | 21.75 (0.50; 315.31) |
| Kruskal–Wallis test: p = 0.3571 | ||||
| VEGFR2, Relative Units | used in calculation | 0.50 (0.13; 1.07) | 1.27 (0.32; 16.00) | 0.91 (0.45; 1.82) *** |
| Kruskal–Wallis test: p = 0.4724 | ||||
| CAIX, Relative Units | used in calculation | 1.21 (0.04; 3.41) | 0.04 (0.01; 1.32) | 1.98 (1.14; 2.28) *** |
| Kruskal–Wallis test: p = 0.3568 | ||||
| Indicator | Localized Kidney Cancer Tissue | Disseminated Kidney Cancer Tissue | RCC Metastatic Tissue |
|---|---|---|---|
| The content of AKT, phospho-PDK1, phospho-c-RAF, phospho-GSK-3beta and phospho-PTEN, m-TOR, phospho-p70 S6 kinase and phospho-4E-BP1 | |||
| phospho -PTEN, % to normal kidney parenchyma | 99.7 (70.4–139.2) | 55.3 (55.2–57.0) * | 50,3 (47.4–194.4) * |
| Kruskal–Wallis test: p = 0.802 | |||
| AKT (total), % to normal kidney parenchyma | 139.95 (110.68–229.4) | 160.35 (112.3–175.4) | 168.4 (113.7–224.1) |
| Kruskal–Wallis test: p = 0.4802 | |||
| phospho -AKT (T308), % to normal kidney parenchyma | 105.1 (76.1–156.58) | 73.1 (55.9–84.7) * | 64.1 (43.1–210.9) * |
| Kruskal–Wallis test: p = 0.03802 | |||
| phospho -AKT (S473), % to normal kidney parenchyma | 118.6 (93.2–164.1) | 85.22 (53.44–106.7) * | 44.1 (20.1–110.9) * |
| Kruskal–Wallis test: p = 0.00802 | |||
| phospho -GSK-3β, % to normal kidney parenchyma | 156.1 (106.85–250.5) | 147.5 (97.4–196.9) | 151.5 (100.1–227.0) |
| Kruskal–Wallis test: p = 0.4802 | |||
| phospho –PDK1, % to normal kidney parenchyma | 132.0 (102.35–168.0) | 119.2 (111.7–132.6) | 150.9 (54.2–170.2) |
| Kruskal–Wallis test: p = 0.1802 | |||
| phospho -c-Raf, % to normal kidney parenchyma | 157.1 (102.9–240.75) | 146.35 (75.8–164.9) | 164.0 (18.1–234.5) |
| Kruskal–Wallis test: p = 0.5802 | |||
| m-TOR, % to normal kidney parenchyma | 158.6 (90.35–218.0) | 129.4 (78.8–138.4) | 124.2 (50.7–194.1) |
| Kruskal–Wallis test: p = 0.2802 | |||
| phospho -mTOR (Ser2448), % to normal kidney parenchyma | 128.1 (93.0–205.6) | 108.1 (81.7–160.4) | 188.8 (72.6–201.7) * |
| Kruskal–Wallis test: p = 0.3802 | |||
| phospho-p70 S6 kinase, % to normal kidney parenchyma | 93.6 (67.1–117.8) | 101.9 (39.0–173.7) | 102.6 (31.2–195.5) |
| Kruskal–Wallis test: p = 0.1641 | |||
| phospho-4E-BP1, % to normal kidney parenchyma | 131.0 (78.1–188.7) | 128.0 (105.4–136.8) | 266.7 (105.5–489.9) |
| Kruskal–Wallis test: p = 0.5284 | |||
| mRNA level of AKT, phospho-PDK1, phospho-c-Raf, phospho-GSK-3beta and phospho-PTEN, m-TOR, 70 S6 kinase and 4E-BP1 | |||
| PTEN, Relative units | 0.80 (0.25; 4.72) | 2.00 (1.10; 8.00) ** | 128.58 (0.25;402.33) *** |
| Kruskal–Wallis test: p = 0.3284 | |||
| AKT, Relative units | 1.19 (0.48; 4.00) | 4.94 (1.74; 23.92) | 0.92 (0.31; 4.75) |
| Kruskal–Wallis test: p = 0.1641 | |||
| GSK-3β, Relative units | 2.41 (0.46; 9.50) | 4.00 (0.26; 16.00) | 0.20 (0.15; 4.12) |
| Kruskal–Wallis test: p = 0.3427 | |||
| PDK, Relative units | 13.58 (1.25; 30.50) | 1.50 (0.03; 8.00) ** | 0.33 (0.15; 1.18) |
| Kruskal–Wallis test: p = 0.0232 | |||
| c-Raf, Relative units | 1.58 (0.11; 6.99) | 2.25 (0.13; 8.00) | 0.15 (0.02; 5.12) |
| Kruskal–Wallis test: p = 0.6720 | |||
| m-TOR, Relative units | 2.69 (0.46; 13.28) | 1.04 (0.34; 8.00) | 0.28 (0.21; 1.15) *** |
| Kruskal–Wallis test: p = 0.3802 | |||
| 70S 6 kinase, Relative units | 1.34 (0.50; 11.45) | 1.69 (0.52; 10.25) | 8.14 (0.06; 72.07) |
| Kruskal–Wallis test: p = 0.9620 | |||
| 4E-BP1, Relative units | 0.83 (0.22; 4.46) | 0.83 (0.25; 7.14) | 0.21 (0.09; 5.40) |
| Kruskal–Wallis test: p = 0.5005 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spirina, L.V.; Yurmazov, Z.A.; Gorbunov, A.K.; Usynin, E.A.; Lushnikova, N.A.; Kovaleva, I.V. Molecular Protein and Expression Profile in the Primary Tumors of Clear Cell Renal Carcinoma and Metastases. Cells 2020, 9, 1680. https://doi.org/10.3390/cells9071680
Spirina LV, Yurmazov ZA, Gorbunov AK, Usynin EA, Lushnikova NA, Kovaleva IV. Molecular Protein and Expression Profile in the Primary Tumors of Clear Cell Renal Carcinoma and Metastases. Cells. 2020; 9(7):1680. https://doi.org/10.3390/cells9071680
Chicago/Turabian StyleSpirina, Liudmila V., Zahar A. Yurmazov, Alexey K. Gorbunov, Evgeny A. Usynin, Nadezhda A. Lushnikova, and Irina V. Kovaleva. 2020. "Molecular Protein and Expression Profile in the Primary Tumors of Clear Cell Renal Carcinoma and Metastases" Cells 9, no. 7: 1680. https://doi.org/10.3390/cells9071680
APA StyleSpirina, L. V., Yurmazov, Z. A., Gorbunov, A. K., Usynin, E. A., Lushnikova, N. A., & Kovaleva, I. V. (2020). Molecular Protein and Expression Profile in the Primary Tumors of Clear Cell Renal Carcinoma and Metastases. Cells, 9(7), 1680. https://doi.org/10.3390/cells9071680