A New Assessment of Thioester-Containing Proteins Diversity of the Freshwater Snail Biomphalaria glabrata
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Statements
2.2. Database Mining, Gene Identification, and Sequence Analysis
2.3. RNA Extraction and Quantitative RT-PCR Analysis
2.4. PCR Analysis of BgTEP2 Isoforms
2.5. Phylogenetic Analyses
3. Results
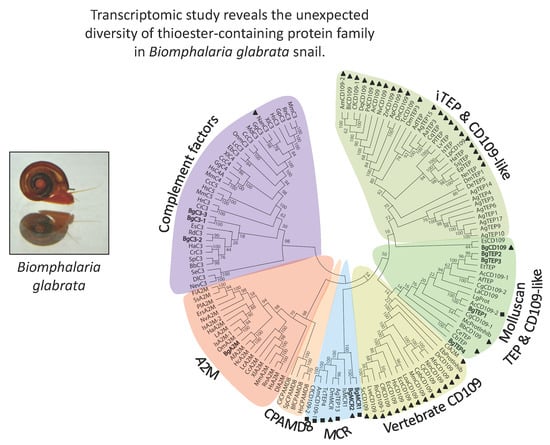
3.1. Phylogenetic Analysis of B. glabrata Thioester-Containing Proteins
3.2. BgTEP Protein Features Analysis
3.3. Organization and Structure of BgTEP Genes
3.4. BgTEPs Genes Expression Pattern in Snail Tissues
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shokal, U.; Eleftherianos, I. Evolution and Function of Thioester-Containing Proteins and the Complement System in the Innate Immune Response. Front. Immunol. 2017, 8, 759. [Google Scholar] [CrossRef]
- Theopold, U.; Schmid, M. Thioester-containing proteins: At the crossroads of immune effector mechanisms. Virulence 2017, 8, 1468–1470. [Google Scholar] [CrossRef] [Green Version]
- Dodds, M.W.; Law, A.S.K. The phylogeny and evolution of the thioester bond-containing proteins C3, C4 and α2–macroglobulin. Immunol. Rev. 1998, 166, 15–26. [Google Scholar] [CrossRef]
- Barrett, A.J.; Starkey, P.M. The interaction of α 2-macroglobulin with proteinases. Characteristics and specificity of the reaction, and a hypothesis concerning its molecular mechanism. Biochem. J. 1973, 133, 709–724. [Google Scholar] [CrossRef]
- Armstrong, P.B. Proteases and protease inhibitors: A balance of activities in host-pathogen interaction. Immunobiology 2006, 211, 263–281. [Google Scholar] [CrossRef]
- Armstrong, P.B.; Quigley, J.P. Alpha2-macroglobulin: An evolutionarily conserved arm of the innate immune system. Dev. Comp. Immunol. 1999, 23, 375–390. [Google Scholar] [CrossRef]
- Garcia-Ferrer, I.; Marrero, A.; Gomis-Ruth, F.X.; Goulas, T. alpha2-Macroglobulins: Structure and Function. Sub-Cell. Biochem. 2017, 83, 149–183. [Google Scholar]
- Rehman, A.A.; Ahsan, H.; Khan, F.H. α-2-Macroglobulin: A physiological guardian. J. Cell. Physiol. 2013, 228, 1665–1675. [Google Scholar] [CrossRef] [PubMed]
- Muller-Eberhard, H.J. Complement. Annu. Rev. Biochem. 1975, 44, 697–724. [Google Scholar] [CrossRef] [PubMed]
- Ricklin, D.; Reis, E.S.; Mastellos, D.C.; Gros, P.; Lambris, J.D. Complement component C3—The “Swiss Army Knife” of innate immunity and host defense. Immunol. Rev. 2016, 274, 33–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricklin, D.; Hajishengallis, G.; Yang, K.; Lambris, J.D. Complement: A key system for immune surveillance and homeostasis. Nat. Immunol. 2010, 11, 785–797. [Google Scholar] [CrossRef] [Green Version]
- Nonaka, M. The complement C3 protein family in invertebrates. Invertebr. Surviv. J. 2011, 8, 21–32. [Google Scholar]
- Bou Aoun, R.; Hetru, C.; Troxler, L.; Doucet, D.; Ferrandon, D.; Matt, N. Analysis of Thioester-Containing Proteins during the Innate Immune Response of Drosophila melanogaster. J. Innate Immun. 2010, 3, 52–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagueux, M.; Perrodou, E.; Levashina, E.A.; Capovilla, M.; Hoffmann, J.A. Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc. Natl. Acad. Sci. USA 2000, 97, 11427–11432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stroschein-Stevenson, S.L.; Foley, E.; O’Farrell, P.H.; Johnson, A.D. Identification of Drosophila Gene Products Required for Phagocytosis of Candida albicans. PLoS Biol. 2006, 4, e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, R.H.G.; Chang, C.-I.; Chelliah, Y.; Levashina, E.A.; Deisenhofer, J. Structural basis for conserved complement factor-like function in the antimalarial protein TEP1. Proc. Natl. Acad. Sci. USA 2007, 104, 11615–11620. [Google Scholar] [CrossRef] [Green Version]
- Buresova, V.; Hajdusek, O.; Franta, Z.; Loosova, G.; Grunclova, L.; Levashina, E.A.; Kopáček, P. Functional Genomics of Tick Thioester-Containing Proteins Reveal the Ancient Origin of the Complement System. J. Innate Immun. 2011, 3, 623–630. [Google Scholar] [CrossRef]
- Castillo, J.C.; Creasy, T.; Kumari, P.; Shetty, A.; Shokal, U.; Tallon, L.J.; Eleftherianos, I. Drosophila anti-nematode and antibacterial immune regulators revealed by RNA-Seq. BMC Genom. 2015, 16, 519. [Google Scholar] [CrossRef] [Green Version]
- Shokal, U.; Kopydlowski, H.; Eleftherianos, I. The distinct function of Tep2 and Tep6 in the immune defense of Drosophila melanogaster against the pathogen Photorhabdus. Virulence 2017, 8, 1668–1682. [Google Scholar] [CrossRef] [Green Version]
- Dostálová, A.; Rommelaere, S.; Poidevin, M.; Lemaitre, B. Thioester-containing proteins regulate the Toll pathway and play a role in Drosophila defence against microbial pathogens and parasitoid wasps. BMC Biol. 2017, 15, 79. [Google Scholar] [CrossRef]
- Blandin, S.; Shiao, S.H.; Moita, L.F.; Janse, C.J.; Waters, A.P.; Kafatos, F.C.; Levashina, E.A. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 2004, 116, 661–670. [Google Scholar] [CrossRef] [Green Version]
- Blandin, S.A.; Marois, E.; Levashina, E.A. Antimalarial Responses in Anopheles gambiae: From a Complement-like Protein to a Complement-like Pathway. Cell Host Microbe 2008, 3, 364–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, G.; Liu, L.; Wang, P.; Zhang, Y.; Zhao, Y.O.; Colpitts, T.M.; Feitosa, F.; Anderson, J.F.; Fikrig, E. An in vivo transfection approach elucidates a role for Aedes aegypti thioester-containing proteins in flaviviral infection. PLoS ONE 2011, 6, e22786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, X.; Liu, Y.; Zhang, X.; Wang, J.; Li, Z.; Pang, X.; Wang, P.; Cheng, G. Complement-related proteins control the flavivirus infection of Aedes aegypti by inducing antimicrobial peptides. PLoS Pathog. 2014, 10, e1004027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yazzie, N.; Salazar, K.A.; Castillo, M.G. Identification, molecular characterization, and gene expression analysis of a CD109 molecule in the Hawaiian bobtail squid Euprymna scolopes. Fish Shellfish Immunol. 2015, 44, 342–355. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Mai, K.; Xu, W.; Liufu, Z. Molecular cloning of alpha2- macroglobulin in sea scallop Chlamys farreri (Bivalvia, Mollusca). Fish Shellfish Immunol. 2005, 18, 345–349. [Google Scholar] [CrossRef]
- Xue, Z.; Wang, L.; Liu, Z.; Wang, W.; Liu, C.; Song, X.; Wang, L.; Song, L. The fragmentation mechanism and immune-protective effect of CfTEP in the scallop Chlamys farreri. Dev. Comp. Immunol. 2017, 76, 220–228. [Google Scholar] [CrossRef]
- Zhang, H.; Song, L.; Li, C.; Zhao, J.; Wang, H.; Gao, Q.; Xu, W. Molecular cloning and characterization of a thioester-containing protein from Zhikong scallop Chlamys farreri. Mol. Immunol. 2007, 44, 3492–3500. [Google Scholar] [CrossRef]
- Gorbushin, A.M. Immune repertoire in the transcriptome of Littorina littorea reveals new trends in lophotrochozoan proto-complement evolution. Dev. Comp. Immunol. 2018, 84, 250–263. [Google Scholar] [CrossRef]
- Mitta, G.; Gourbal, B.; Grunau, C.; Knight, M.; Bridger, J.M.; Theron, A. The Compatibility Between Biomphalaria glabrata Snails and Schistosoma mansoni: An Increasingly Complex Puzzle. Adv. Parasitol. 2016, 97, 111–145. [Google Scholar]
- Pila, E.A.; Li, H.; Hambrook, J.R.; Wu, X.; Hanington, P.C. Schistosomiasis from a Snail’s Perspective: Advances in Snail Immunity. Trends Parasitol. 2017, 33, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Portet, A.; Pinaud, S.; Tetreau, G.; Galinier, R.; Cosseau, C.; Duval, D.; Grunau, C.; Mitta, G.; Gourbal, B. Integrated multi-omic analyses in Biomphalaria-Schistosoma dialogue reveal the immunobiological significance of FREP-SmPoMuc interaction. Dev. Comp. Immunol. 2017, 75, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Bouchut, A.; Roger, E.; Gourbal, B.; Grunau, C.; Coustau, C.; Mitta, G. The compatibiuty polymorphism in invertebrate host/trematodes interactions: Research of molecular determinants. Parasite 2008, 15, 304–309. [Google Scholar] [CrossRef] [Green Version]
- Mitta, G.; Adema, C.M.; Gourbal, B.; Loker, E.S.; Theron, A. Compatibility polymorphism in snail/schistosome interactions: From field to theory to molecular mechanisms. Dev. Comp. Immunol. 2012, 37, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theron, A.; Coustau, C. Are Biomphalaria snails resistant to Schistosoma mansoni? J. Helminthol. 2005, 79, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Morand, S.; Manning, S.D.; Woolhouse, M.E. Parasite-host coevolution and geographic patterns of parasite infectivity and host susceptibility. Proceedings 1996, 263, 119–128. [Google Scholar]
- Combes, C. Selective pressure in host-parasite systems. J. Soc. Biol. 2000, 194, 19–23. [Google Scholar] [CrossRef]
- Galinier, R.; Portela, J.; Moné, Y.; Allienne, J.F.; Henri, H.; Delbecq, S.; Mitta, G.; Gourbal, B.; Duval, D. Biomphalysin, a New β Pore-forming Toxin Involved in Biomphalaria glabrata Immune Defense against Schistosoma mansoni. PLoS Pathog. 2013, 9, e1003216. [Google Scholar] [CrossRef]
- Moné, Y.; Gourbal, B.; Duval, D.; du Pasquier, L.; Kieffer-Jaquinod, S.; Mitta, G. A large repertoire of parasite epitopes matched by a large repertoire of host immune receptors in an invertebrate host/parasite model. PLoS Negl. Trop. Dis. 2010, 4, e813. [Google Scholar] [CrossRef] [Green Version]
- Adema, C.M.; Hertel, L.A.; Miller, R.D.; Loker, E.S. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc. Natl. Acad. Sci. USA 1997, 94, 8691–8696. [Google Scholar] [CrossRef] [Green Version]
- Roger, E.; Grunau, C.; Pierce, R.J.; Hirai, H.; Gourbal, B.; Galinier, R.; Emans, R.; Cesari, I.M.; Cosseau, C.; Mitta, G. Controlled Chaos of Polymorphic Mucins in a Metazoan Parasite (Schistosoma mansoni) Interacting with Its Invertebrate Host (Biomphalaria glabrata). PLoS Negl. Trop. Dis. 2008, 2, e330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galinier, R.; Roger, E.; Mone, Y.; Duval, D.; Portet, A.; Pinaud, S.; Chaparro, C.; Grunau, C.; Genthon, C.; Dubois, E.; et al. A multistrain approach to studying the mechanisms underlying compatibility in the interaction between Biomphalaria glabrata and Schistosoma mansoni. PLoS Negl. Trop. Dis. 2017, 11, e0005398. [Google Scholar] [CrossRef] [PubMed]
- Mone, Y.; Ribou, A.C.; Cosseau, C.; Duval, D.; Theron, A.; Mitta, G.; Gourbal, B. An example of molecular co-evolution: Reactive oxygen species (ROS) and ROS scavenger levels in Schistosoma mansoni/Biomphalaria glabrata interactions. Int. J. Parasitol. 2011, 41, 721–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.J.; Dinguirard, N.; Sabat, G.; Lui, H.D.; Gonzalez, L.; Gehring, M.; Bickham-Wright, U.; Yoshino, T.P. Proteomic analysis of Biomphalaria glabrata plasma proteins with binding affinity to those expressed by early developing larval Schistosoma mansoni. PLoS Pathog. 2017, 13, e1006081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portet, A.; Galinier, R.; Pinaud, S.; Portela, J.; Nowacki, F.; Gourbal, B.; Duval, D. BgTEP: An Antiprotease Involved in Innate Immune Sensing in Biomphalaria glabrata. Front. Immunol. 2018, 9, 1206. [Google Scholar] [CrossRef] [Green Version]
- Bender, R.C.; Bayne, C.J. Purification and characterization of a tetrameric α-macroglobulin proteinase inhibitor from the gastropod mollusc Biomphalaria glabrata. Biochem. J. 1996, 316 Pt 3, 893–900. [Google Scholar] [CrossRef] [Green Version]
- Bender, R.C.; Fryer, S.E.; Bayne, C.J. Proteinase inhibitory activity in the plasma of a mollusc: Evidence for the presence of α-macroglobulin in Biomphalaria glabrata. Comp. Biochem. Physiol. B Comp. Biochem. 1992, 102, 821–824. [Google Scholar] [CrossRef]
- Fryer, S.E.; Bender, R.C.; Bayne, C.J. Inhibition of cysteine proteinase from Schistosoma mansoni larvae by α-macroglobulin from the plasma of Biomphalaria glabrata. J. Parasitol. 1996, 82, 343–347. [Google Scholar] [CrossRef]
- Adema, C.M.; Hillier, L.W.; Jones, C.S.; Loker, E.S.; Knight, M.; Minx, P.; Oliveira, G.; Raghavan, N.; Shedlock, A.; do Amaral, L.R.; et al. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat. Commun. 2017, 8, 15451. [Google Scholar] [CrossRef]
- Dheilly, N.M.; Duval, D.; Mouahid, G.; Emans, R.; Allienne, J.-F.; Galinier, R.; Genthon, C.; Dubois, E.; Du Pasquier, L.; Adema, C.M.; et al. A family of variable immunoglobulin and lectin domain containing molecules in the snail Biomphalaria glabrata. Dev. Comp. Immunol. 2015, 48, 234–243. [Google Scholar] [CrossRef] [Green Version]
- Pinaud, S.; Portela, J.; Duval, D.; Nowacki, F.C.; Olive, M.A.; Allienne, J.F.; Galinier, R.; Dheilly, N.M.; Kieffer-Jaquinod, S.; Mitta, G.; et al. A Shift from Cellular to Humoral Responses Contributes to Innate Immune Memory in the Vector Snail Biomphalaria glabrata. PLoS Pathog. 2016, 12, e1005361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef] [PubMed]
- Penn, O.; Privman, E.; Ashkenazy, H.; Landan, G.; Graur, D.; Pupko, T. GUIDANCE: A web server for assessing alignment confidence scores. Nucleic Acids Res. 2010, 38, W23–W28. [Google Scholar] [CrossRef] [Green Version]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Sottrup-Jensen, L.; Sand, O.; Kristensen, L.; Fey, G.H. The α-macroglobulin bait region. Sequence diversity and localization of cleavage sites for proteinases in five mammalian α-macroglobulins. J. Biol. Chem. 1989, 264, 15781–15789. [Google Scholar]
- Banyai, L.; Patthy, L. The NTR module: Domains of netrins, secreted frizzled related proteins, and type I procollagen C-proteinase enhancer protein are homologous with tissue inhibitors of metalloproteases. Protein Sci. 1999, 8, 1636–1642. [Google Scholar] [CrossRef] [Green Version]
- de Bruijn, M.H.; Fey, G.H. Human complement component C3: cDNA coding sequence and derived primary structure. Proc. Natl. Acad. Sci. USA 1985, 82, 708–712. [Google Scholar] [CrossRef] [Green Version]
- Gerard, N.P.; Lively, M.O.; Gerard, C. Amino acid sequence of guinea pig C3a anaphylatoxin. Protein Seq. Data Anal. 1988, 1, 473–478. [Google Scholar]
- Prosper, J.Y.A. Characterization of CD109. In Graduate Department of Medical Biophysics; University of Toronto: Toronto, ON, Canada, 2011. [Google Scholar]
- Law, S.K.; Dodds, A.W. The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 1997, 6, 263–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gadjeva, M.; Dodds, A.W.; Taniguchi-Sidle, A.; Willis, A.C.; Isenman, D.E.; Law, S.K. The covalent binding reaction of complement component C3. J. Immunol. 1998, 161, 985–990. [Google Scholar] [PubMed]
- Ma, H.; Wang, B.; Zhang, J.; Li, F.; Xiang, J. Multiple forms of α-2 macroglobulin in shrimp Fenneropenaeus chinesis and their transcriptional response to WSSV or Vibrio pathogen infection. Dev. Comp. Immunol. 2010, 34, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F.; et al. The genome sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holt, R.A.; Subramanian, G.M.; Halpern, A.; Sutton, G.G.; Charlab, R.; Nusskern, D.R.; Wincker, P.; Clark, A.G.; Ribeiro, J.M.; Wides, R.; et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science 2002, 298, 129–149. [Google Scholar] [CrossRef] [PubMed]
- Falade, M.O.; Otarigho, B. Comparative Functional Study of Thioester-containing Related Proteins in the Recently Sequenced Genome of Biomphalaria glabrata. Iran. J. Parasitol. 2018, 13, 79–88. [Google Scholar]
- Liao, H.; Wang, J.; Xun, X.; Zhao, L.; Yang, Z.; Zhu, X.; Xing, Q.; Huang, X.; Bao, Z. Identification and characterization of TEP family genes in Yesso scallop (Patinopecten yessoensis) and their diverse expression patterns in response to bacterial infection. Fish Shellfish Immunol. 2018, 79, 327–339. [Google Scholar] [CrossRef]
- Matetovici, I.; Van Den Abbeele, J. Thioester-containing proteins in the tsetse fly (Glossina) and their response to trypanosome infection. Insect Mol. Biol. 2018, 27, 414–428. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, K.L.; Sottrup-Jensen, L.; Nagase, H.; Thogersen, H.C.; Etzerodt, M. Amino acid sequence of hen ovomacroglobulin (ovostatin) deduced from cloned cDNA. DNA Seq. 1994, 5, 111–119. [Google Scholar] [CrossRef]
- Nagase, H.; Harris, E.D., Jr. Ovostatin: A novel proteinase inhibitor from chicken egg white. II. Mechanism of inhibition studied with collagenase and thermolysin. J. Biol. Chem. 1983, 258, 7490–7498. [Google Scholar]
- Levashina, E.A.; Moita, L.F.; Blandin, S.; Vriend, G.; Lagueux, M.; Kafatos, F.C. Conserved Role of a Complement-like Protein in Phagocytosis Revealed by dsRNA Knockout in Cultured Cells of the Mosquito, Anopheles gambiae. Cell 2001, 104, 709–718. [Google Scholar] [CrossRef]
- Batz, T.; Forster, D.; Luschnig, S. The transmembrane protein Macroglobulin complement-related is essential for septate junction formation and epithelial barrier function in Drosophila. Development 2014, 141, 899–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.; Rodrigues, F.; Kary, C.; Contet, A.; Logan, M.; Baxter, R.H.G.; Wood, W.; Baehrecke, E.H. Complement-Related Regulates Autophagy in Neighboring Cells. Cell 2017, 170, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Shokal, U.; Kopydlowski, H.; Harsh, S.; Eleftherianos, I. Thioester-Containing Proteins 2 and 4 Affect the Metabolic Activity and Inflammation Response in Drosophila. Infect. Immun. 2018, 86, e00810-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pompon, J.; Levashina, E.A. A New Role of the Mosquito Complement-like Cascade in Male Fertility in Anopheles gambiae. PLoS Biol. 2015, 13, e1002255. [Google Scholar] [CrossRef] [Green Version]
- Guillou, F.; Mitta, G.; Galinier, R.; Coustau, C. Identification and expression of gene transcripts generated during an anti-parasitic response in Biomphalaria glabrata. Dev. Comp. Immunol. 2007, 31, 657–671. [Google Scholar] [CrossRef]
- Lin, M.; Sutherland, D.R.; Horsfall, W.; Totty, N.; Yeo, E.; Nayar, R.; Wu, X.F.; Schuh, A.C. Cell surface antigen CD109 is a novel member of the α(2) macroglobulin/C3, C4, C5 family of thioester-containing proteins. Blood 2002, 99, 1683–1691. [Google Scholar] [CrossRef]
- Finnson, K.W.; Tam, B.Y.; Liu, K.; Marcoux, A.; Lepage, P.; Roy, S.; Bizet, A.A.; Philip, A. Identification of CD109 as part of the TGF-β receptor system in human keratinocytes. FASEB J. 2006, 20, 1525–1527. [Google Scholar] [CrossRef]
- Hagiwara, S.; Murakumo, Y.; Sato, T.; Shigetomi, T.; Mitsudo, K.; Tohnai, I.; Ueda, M.; Takahashi, M. Up-regulation of CD109 expression is associated with carcinogenesis of the squamous epithelium of the oral cavity. Cancer Sci. 2008, 99, 1916–1923. [Google Scholar] [CrossRef]
- Mii, S.; Hoshino, A.; Enomoto, A.; Murakumo, Y.; Ito, M.; Yamaguchi, A.; Takahashi, M. CD109 deficiency induces osteopenia with an osteoporosis-like phenotype in vivo. Genes Cells 2018, 23, 590–598. [Google Scholar] [CrossRef]
- Zhou, S.; da Silva, S.D.; Siegel, P.M.; Philip, A. CD109 acts as a gatekeeper of the epithelial trait by suppressing epithelial to mesenchymal transition in squamous cell carcinoma cells in vitro. Sci. Rep. 2019, 9, 16317. [Google Scholar] [CrossRef] [Green Version]
- Collins, A.J.; Schleicher, T.R.; Rader, B.A.; Nyholm, S.V. Understanding the role of host hemocytes in a squid/vibrio symbiosis using transcriptomics and proteomics. Front. Immunol. 2012, 3, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Lookeren Campagne, M.; Wiesmann, C.; Brown, E.J. Macrophage complement receptors and pathogen clearance. Cell. Microbiol. 2007, 9, 2095–2102. [Google Scholar] [CrossRef] [PubMed]
- Alcorlo, M.; Lopez-Perrote, A.; Delgado, S.; Yebenes, H.; Subias, M.; Rodriguez-Gallego, C.; Rodriguez de Cordoba, S.; Llorca, O. Structural insights on complement activation. FEBS J. 2015, 282, 3883–3891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, M.; Contet, A.; Hou, C.D.; Levashina, E.A.; Baxter, R.H.G. Anopheles gambiae TEP1 forms a complex with the coiled-coil domain of LRIM1/APL1C following a conformational change in the thioester domain. PLoS ONE 2019, 14, e0218203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Wang, L.; Song, L.; Zhao, J.; Qiu, L.; Gao, Y.; Song, X.; Li, L.; Zhang, Y.; Zhang, L. The genomic structure, alternative splicing and immune response of Chlamys farreri thioester-containing protein. Dev. Comp. Immunol. 2009, 33, 1070–1076. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duval, D.; Pichon, R.; Lassalle, D.; Laffitte, M.; Gourbal, B.; Galinier, R. A New Assessment of Thioester-Containing Proteins Diversity of the Freshwater Snail Biomphalaria glabrata. Genes 2020, 11, 69. https://doi.org/10.3390/genes11010069
Duval D, Pichon R, Lassalle D, Laffitte M, Gourbal B, Galinier R. A New Assessment of Thioester-Containing Proteins Diversity of the Freshwater Snail Biomphalaria glabrata. Genes. 2020; 11(1):69. https://doi.org/10.3390/genes11010069
Chicago/Turabian StyleDuval, David, Remi Pichon, Damien Lassalle, Maud Laffitte, Benjamin Gourbal, and Richard Galinier. 2020. "A New Assessment of Thioester-Containing Proteins Diversity of the Freshwater Snail Biomphalaria glabrata" Genes 11, no. 1: 69. https://doi.org/10.3390/genes11010069
APA StyleDuval, D., Pichon, R., Lassalle, D., Laffitte, M., Gourbal, B., & Galinier, R. (2020). A New Assessment of Thioester-Containing Proteins Diversity of the Freshwater Snail Biomphalaria glabrata. Genes, 11(1), 69. https://doi.org/10.3390/genes11010069