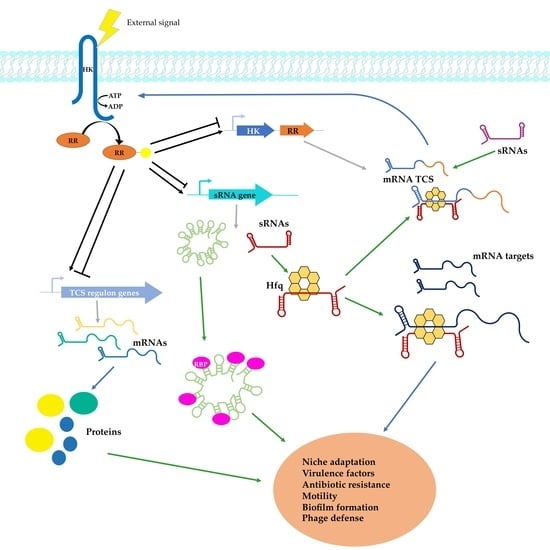
Interplay between Regulatory RNAs and Signal Transduction Systems during Bacterial Infection
Abstract
:1. Introduction
2. sRNAs Regulating Host-Pathogen Interactions
2.1. sRNAs Involved in Stress Response
2.1.1. sRNAs Regulated by Alternative σ-Factors
2.1.2. Other ESR-Regulated sRNAs
2.2. TCS-Associated sRNAs Involved in Quorum-Sensing Signaling
2.3. TCS-Associated sRNAs Involved in CRISPR-Cas Immunity and Competence Control
2.4. Signal Transduction Systems-Associated Riboswitches
2.5. Other TCS-Associated sRNAs Involved in Virulence
3. Participation of sRNAs in Regulatory Networks
4. Strategies to Identify TCS-Associated sRNAs
4.1. Global Approaches for sRNA Identification
4.2. Specific Approaches Adapted for TCS-Associated sRNAs
5. Antimicrobial Strategies Related with TCS and Associated sRNAs
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mitchell, A.; Romano, G.H.; Groisman, B.; Yona, A.; Dekel, E.; Kupiec, M.; Dahan, O.; Pilpel, Y. Adaptive prediction of environmental changes by microorganisms. Nature 2009, 460, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bleuven, C.; Landry, C.R. Molecular and cellular bases of adaptation to a changing environment in microorganisms. Proc. Biol. Sci. 2016, 283. [Google Scholar] [CrossRef]
- Wagner, E.G.H.; Romby, P. Small RNAs in bacteria and archaea: Who they are, what they do, and how they do it. Adv. Genet. 2015, 90, 133–208. [Google Scholar] [CrossRef] [PubMed]
- Gorski, S.A.; Vogel, J.; Doudna, J.A. RNA-based recognition and targeting: Sowing the seeds of specificity. Nat. Rev. Mol. Cell Biol. 2017, 18, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA Translation and Stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waters, L.S.; Storz, G. Regulatory RNAs in bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliva, G.; Sahr, T.; Buchrieser, C. Small RNAs, 5′ UTR elements and RNA-binding proteins in intracellular bacteria: Impact on metabolism and virulence. FEMS Microbiol. Rev. 2015, 39, 331–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soutourina, O. RNA-based control mechanisms of Clostridium difficile. Curr. Opin. Microbiol. 2017, 36, 62–68. [Google Scholar] [CrossRef]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef] [Green Version]
- Smirnov, A.; Wang, C.; Drewry, L.L.; Vogel, J. Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J. 2017, 36, 1029–1045. [Google Scholar] [CrossRef]
- Romby, P.; Charpentier, E. An overview of RNAs with regulatory functions in gram-positive bacteria. Cell. Mol. Life Sci. 2010, 67, 217–237. [Google Scholar] [CrossRef]
- Brosse, A.; Guillier, M. Bacterial Small RNAs in Mixed Regulatory Networks. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Göpel, Y.; Görke, B. Rewiring two-component signal transduction with small RNAs. Curr. Opin. Microbiol. 2012, 15, 132–139. [Google Scholar] [CrossRef]
- Mandin, P.; Guillier, M. Expanding control in bacteria: Interplay between small RNAs and transcriptional regulators to control gene expression. Curr. Opin. Microbiol. 2013, 16, 125–132. [Google Scholar] [CrossRef]
- Chakravarty, S.; Massé, E. RNA-Dependent Regulation of Virulence in Pathogenic Bacteria. Front. Cell. Infect. Microbiol. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Hews, C.L.; Cho, T.; Rowley, G.; Raivio, T.L. Maintaining Integrity Under Stress: Envelope Stress Response Regulation of Pathogenesis in Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Pucciarelli, M.G.; García-del Portillo, F. Within-Host Envelope Remodelling and its Impact in Bacterial Pathogen Recognition. Curr. Issues Mol. Biol. 2018, 25, 43–60. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Klein, G.; Raina, S. Small regulatory bacterial RNAs regulating the envelope stress response. Biochem. Soc. Trans. 2017, 45, 417–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kill, K.; Binnewies, T.T.; Sicheritz-Pontén, T.; Willenbrock, H.; Hallin, P.F.; Wassenaar, T.M.; Ussery, D.W. Genome update: Sigma factors in 240 bacterial genomes. Microbiology 2005, 151, 3147–3150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, R.; Stock, A.M. Biological Insights from Structures of Two-Component Proteins. Annu. Rev. Microbiol. 2009, 63, 133–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groisman, E.A. Feedback Control of Two-Component Regulatory Systems. Annu. Rev. Microbiol. 2016, 70, 103–124. [Google Scholar] [CrossRef]
- Alm, E.; Huang, K.; Arkin, A. The Evolution of Two-Component Systems in Bacteria Reveals Different Strategies for Niche Adaptation. PLoS Comput. Biol. 2006, 2. [Google Scholar] [CrossRef] [PubMed]
- Grabowicz, M.; Silhavy, T.J. Envelope stress responses: An interconnected safety net. Trends Biochem. Sci. 2017, 42, 232–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, Y.; Vogel, J. A 3′ UTR-Derived Small RNA Provides the Regulatory Noncoding Arm of the Inner Membrane Stress Response. Mol. Cell 2016, 61, 352–363. [Google Scholar] [CrossRef] [Green Version]
- Grabowicz, M.; Koren, D.; Silhavy, T.J. The CpxQ sRNA Negatively Regulates Skp to Prevent Mistargeting of β-Barrel Outer Membrane Proteins into the Cytoplasmic Membrane. mBio 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Vogt, S.L.; Evans, A.D.; Guest, R.L.; Raivio, T.L. The Cpx Envelope Stress Response Regulates and Is Regulated by Small Noncoding RNAs. J. Bacteriol. 2014, 196, 4229–4238. [Google Scholar] [CrossRef] [Green Version]
- De Lay, N.; Gottesman, S. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 2009, 191, 461–476. [Google Scholar] [CrossRef] [Green Version]
- Lalaouna, D.; Prévost, K.; Laliberté, G.; Houé, V.; Massé, E. Contrasting silencing mechanisms of the same target mRNA by two regulatory RNAs in Escherichia coli. Nucleic Acids Res. 2018, 46, 2600–2612. [Google Scholar] [CrossRef] [Green Version]
- Guillier, M.; Gottesman, S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006, 59, 231–247. [Google Scholar] [CrossRef]
- Guillier, M.; Gottesman, S. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 2008, 36, 6781–6794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmqvist, E.; Reimegård, J.; Sterk, M.; Grantcharova, N.; Römling, U.; Wagner, E.G.H. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010, 29, 1840–1850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Lay, N.; Gottesman, S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 2012, 86, 524–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brosse, A.; Korobeinikova, A.; Gottesman, S.; Guillier, M. Unexpected properties of sRNA promoters allow feedback control via regulation of a two-component system. Nucleic Acids Res. 2016, 44, 9650–9666. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.; Delihas, N. micF RNA binds to the 5’ end of ompF mRNA and to a protein from Escherichia coli. Biochemistry 1990, 29, 9249–9256. [Google Scholar] [CrossRef]
- Holmqvist, E.; Unoson, C.; Reimegård, J.; Wagner, E.G.H. A mixed double negative feedback loop between the sRNA MicF and the global regulator Lrp. Mol. Microbiol. 2012, 84, 414–427. [Google Scholar] [CrossRef]
- Moon, K.; Gottesman, S. A PhoQ/P-Regulated small RNA Regulates Sensitivity of Escherichia coli to Antimicrobial Peptides. Mol. Microbiol. 2009, 74, 1314–1330. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-J.; Gottesman, S. sRNA roles in regulating transcriptional regulators: Lrp and SoxS regulation by sRNAs. Nucleic Acids Res. 2016, 44, 6907–6923. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.-J.; Groisman, E.A. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol. Microbiol. 2010, 76, 1020–1033. [Google Scholar] [CrossRef] [Green Version]
- Westermann, A.J.; Förstner, K.U.; Amman, F.; Barquist, L.; Chao, Y.; Schulte, L.N.; Müller, L.; Reinhardt, R.; Stadler, P.F.; Vogel, J. Dual RNA-seq unveils noncoding RNA functions in host–pathogen interactions. Nature 2016, 529, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Palmer, A.D.; Vanderpool, C.K.; Slauch, J.M. The Small RNA PinT Contributes to PhoP-Mediated Regulation of the Salmonella Pathogenicity Island 1 Type III Secretion System in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, A.D.; Kim, K.; Slauch, J.M. PhoP-Mediated Repression of the SPI1 Type 3 Secretion System in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majdalani, N.; Chen, S.; Murrow, J.; St John, K.; Gottesman, S. Regulation of RpoS by a novel small RNA: The characterization of RprA. Mol. Microbiol. 2001, 39, 1382–1394. [Google Scholar] [CrossRef]
- Guo, X.-P.; Sun, Y.-C. New Insights into the Non-orthodox Two Component Rcs Phosphorelay System. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Latasa, C.; García, B.; Echeverz, M.; Toledo-Arana, A.; Valle, J.; Campoy, S.; García-del Portillo, F.; Solano, C.; Lasa, I. Salmonella Biofilm Development Depends on the Phosphorylation Status of RcsB. J. Bacteriol. 2012, 194, 3708–3722. [Google Scholar] [CrossRef] [Green Version]
- Urban, J.H.; Vogel, J. Two Seemingly Homologous Noncoding RNAs Act Hierarchically to Activate glmS mRNA Translation. PLoS Biol. 2008, 6, e64. [Google Scholar] [CrossRef]
- Reichenbach, B.; Göpel, Y.; Görke, B. Dual control by perfectly overlapping sigma 54- and sigma 70- promoters adjusts small RNA GlmY expression to different environmental signals. Mol. Microbiol. 2009, 74, 1054–1070. [Google Scholar] [CrossRef]
- Gruber, C.C.; Sperandio, V. Global Analysis of Posttranscriptional Regulation by GlmY and GlmZ in Enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2015, 83, 1286–1295. [Google Scholar] [CrossRef] [Green Version]
- Reading, N.C.; Rasko, D.A.; Torres, A.G.; Sperandio, V. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 5889–5894. [Google Scholar] [CrossRef] [Green Version]
- Gruber, C.C.; Sperandio, V. Posttranscriptional control of microbe-induced rearrangement of host cell actin. mBio 2014, 5, e01025-01013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandin, P.; Gottesman, S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010, 29, 3094–3107. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.; Feng, J.; Sachs, G. Helicobacter pylori 5′ureB-sRNA, a cis-Encoded Antisense Small RNA, Negatively Regulates ureAB Expression by Transcription Termination. J. Bacteriol. 2013, 195, 444–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bejerano-Sagie, M.; Xavier, K.B. The role of small RNAs in quorum sensing. Curr. Opin. Microbiol. 2007, 10, 189–198. [Google Scholar] [CrossRef]
- Lenz, D.H.; Mok, K.C.; Lilley, B.N.; Kulkarni, R.V.; Wingreen, N.S.; Bassler, B.L. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 2004, 118, 69–82. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Bassler, B.L. Quorum regulatory small RNAs repress type VI secretion in Vibrio cholerae. Mol. Microbiol. 2014, 92, 921–930. [Google Scholar] [CrossRef] [Green Version]
- Rutherford, S.T.; van Kessel, J.C.; Shao, Y.; Bassler, B.L. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 2011, 25, 397–408. [Google Scholar] [CrossRef] [Green Version]
- Xi, D.; Li, Y.; Yan, J.; Li, Y.; Wang, X.; Cao, B. Small RNA coaR contributes to intestinal colonization in Vibrio cholerae via the two-component system EnvZ/OmpR. Environ. Microbiol. 2019. [Google Scholar] [CrossRef]
- Peterson, K.M.; Gellings, P.S. Multiple intraintestinal signals coordinate the regulation of Vibrio cholerae virulence determinants. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef]
- Sonnleitner, E.; Abdou, L.; Haas, D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2009, 106, 21866–21871. [Google Scholar] [CrossRef] [PubMed]
- Linares, J.F.; Moreno, R.; Fajardo, A.; Martínez-Solano, L.; Escalante, R.; Rojo, F.; Martínez, J.L. The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ. Microbiol. 2010, 12, 3196–3212. [Google Scholar] [CrossRef] [PubMed]
- Wenner, N.; Maes, A.; Cotado-Sampayo, M.; Lapouge, K. NrsZ: A novel, processed, nitrogen-dependent, small non-coding RNA that regulates Pseudomonas aeruginosa PAO1 virulence. Environ. Microbiol. 2014, 16, 1053–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tata, M.; Amman, F.; Pawar, V.; Wolfinger, M.T.; Weiss, S.; Häussler, S.; Bläsi, U. The Anaerobically Induced sRNA PaiI Affects Denitrification in Pseudomonas aeruginosa PA14. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Borges, A.L.; Castro, B.; Govindarajan, S.; Solvik, T.; Escalante, V.; Bondy-Denomy, J. Bacterial alginate regulators and phage homologs repress CRISPR–Cas immunity. Nat. Microbiol. 2020, 5, 679–687. [Google Scholar] [CrossRef]
- Liu, W.; Li, M.; Jiao, L.; Wang, P.; Yan, Y. PmrA/PmrB Two-Component System Regulation of lipA Expression in Pseudomonas aeruginosa PAO1. Front. Microbiol. 2017, 8, 2690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zere, T.R.; Vakulskas, C.A.; Leng, Y.; Pannuri, A.; Potts, A.H.; Dias, R.; Tang, D.; Kolaczkowski, B.; Georgellis, D.; Ahmer, B.M.M.; et al. Genomic Targets and Features of BarA-UvrY (-SirA) Signal Transduction Systems. PLoS ONE 2015, 10, e0145035. [Google Scholar] [CrossRef] [Green Version]
- Chavez, R.G.; Alvarez, A.F.; Romeo, T.; Georgellis, D. The physiological stimulus for the BarA sensor kinase. J. Bacteriol. 2010, 192, 2009–2012. [Google Scholar] [CrossRef] [Green Version]
- Brencic, A.; McFarland, K.A.; McManus, H.R.; Castang, S.; Mogno, I.; Dove, S.L.; Lory, S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 2009, 73, 434–445. [Google Scholar] [CrossRef] [Green Version]
- González, N.; Heeb, S.; Valverde, C.; Kay, E.; Reimmann, C.; Junier, T.; Haas, D. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics 2008, 9, 167. [Google Scholar] [CrossRef] [Green Version]
- Sahr, T.; Brüggemann, H.; Jules, M.; Lomma, M.; Albert-Weissenberger, C.; Cazalet, C.; Buchrieser, C. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. 2009, 72, 741–762. [Google Scholar] [CrossRef] [Green Version]
- Lenz, D.H.; Miller, M.B.; Zhu, J.; Kulkarni, R.V.; Bassler, B.L. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 2005, 58, 1186–1202. [Google Scholar] [CrossRef]
- Sievers, S.; Sternkopf Lillebæk, E.M.; Jacobsen, K.; Lund, A.; Mollerup, M.S.; Nielsen, P.K.; Kallipolitis, B.H. A multicopy sRNA of Listeria monocytogenes regulates expression of the virulence adhesin LapB. Nucleic Acids Res. 2014, 42, 9383–9398. [Google Scholar] [CrossRef] [Green Version]
- Reis, O.; Sousa, S.; Camejo, A.; Villiers, V.; Gouin, E.; Cossart, P.; Cabanes, D. LapB, a novel Listeria monocytogenes LPXTG surface adhesin, required for entry into eukaryotic cells and virulence. J. Infect. Dis. 2010, 202, 551–562. [Google Scholar] [CrossRef] [Green Version]
- Ross, J.A.; Thorsing, M.; Lillebæk, E.M.S.; Teixeira Dos Santos, P.; Kallipolitis, B.H. The LhrC sRNAs control expression of T cell-stimulating antigen TcsA in Listeria monocytogenes by decreasing tcsA mRNA stability. RNA Biol. 2019, 16, 270–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, S.; Lund, A.; Menendez-Gil, P.; Nielsen, A.; Storm Mollerup, M.; Lambert Nielsen, S.; Buch Larsson, P.; Borch-Jensen, J.; Johansson, J.; Kallipolitis, B.H. The multicopy sRNA LhrC controls expression of the oligopeptide-binding protein OppA in Listeria monocytogenes. RNA Biol. 2015, 12, 985–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, P.T.; Menendez-Gil, P.; Sabharwal, D.; Christensen, J.-H.; Brunhede, M.Z.; Lillebæk, E.M.S.; Kallipolitis, B.H. The Small Regulatory RNAs LhrC1-5 Contribute to the Response of Listeria monocytogenes to Heme Toxicity. Front. Microbiol. 2018, 9, 599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mollerup, M.S.; Ross, J.A.; Helfer, A.-C.; Meistrup, K.; Romby, P.; Kallipolitis, B.H. Two novel members of the LhrC family of small RNAs in Listeria monocytogenes with overlapping regulatory functions but distinctive expression profiles. RNA Biol. 2016, 13, 895–915. [Google Scholar] [CrossRef] [Green Version]
- Grubaugh, D.; Regeimbal, J.M.; Ghosh, P.; Zhou, Y.; Lauer, P.; Dubensky, T.W.; Higgins, D.E. The VirAB ABC Transporter Is Required for VirR Regulation of Listeria monocytogenes Virulence and Resistance to Nisin. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frantz, R.; Teubner, L.; Schultze, T.; La Pietra, L.; Müller, C.; Gwozdzinski, K.; Pillich, H.; Hain, T.; Weber-Gerlach, M.; Panagiotidis, G.-D.; et al. The secRNome of Listeria monocytogenes Harbors Small Noncoding RNAs That Are Potent Inducers of Beta Interferon. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solans, L.; Gonzalo-Asensio, J.; Sala, C.; Benjak, A.; Uplekar, S.; Rougemont, J.; Guilhot, C.; Malaga, W.; Martín, C.; Cole, S.T. The PhoP-Dependent ncRNA Mcr7 Modulates the TAT Secretion System in Mycobacterium tuberculosis. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef]
- Novick, R.P.; Ross, H.F.; Projan, S.J.; Kornblum, J.; Kreiswirth, B.; Moghazeh, S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993, 12, 3967–3975. [Google Scholar] [CrossRef]
- Bronesky, D.; Wu, Z.; Marzi, S.; Walter, P.; Geissmann, T.; Moreau, K.; Vandenesch, F.; Caldelari, I.; Romby, P. Staphylococcus aureus RNAIII and Its Regulon Link Quorum Sensing, Stress Responses, Metabolic Adaptation, and Regulation of Virulence Gene Expression. Annu. Rev. Microbiol. 2016, 70, 299–316. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Zhang, X.; Sun, H.; Sun, B. ArtR, a novel sRNA of Staphylococcus aureus, regulates α-toxin expression by targeting the 5’ UTR of sarT mRNA. Med. Microbiol. Immunol. 2014, 203, 1–12. [Google Scholar] [CrossRef]
- Durand, S.; Braun, F.; Lioliou, E.; Romilly, C.; Helfer, A.-C.; Kuhn, L.; Quittot, N.; Nicolas, P.; Romby, P.; Condon, C. A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis in Bacillus subtilis. PLoS Genet. 2015, 11, e1004957. [Google Scholar] [CrossRef] [Green Version]
- Bohn, C.; Rigoulay, C.; Chabelskaya, S.; Sharma, C.M.; Marchais, A.; Skorski, P.; Borezée-Durant, E.; Barbet, R.; Jacquet, E.; Jacq, A.; et al. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010, 38, 6620–6636. [Google Scholar] [CrossRef] [PubMed]
- Schoenfelder, S.M.K.; Lange, C.; Prakash, S.A.; Marincola, G.; Lerch, M.F.; Wencker, F.D.R.; Förstner, K.U.; Sharma, C.M.; Ziebuhr, W. The small non-coding RNA RsaE influences extracellular matrix composition in Staphylococcus epidermidis biofilm communities. PLoS Pathog. 2019, 15, e1007618. [Google Scholar] [CrossRef] [Green Version]
- Rochat, T.; Bohn, C.; Morvan, C.; Le Lam, T.N.; Razvi, F.; Pain, A.; Toffano-Nioche, C.; Ponien, P.; Jacq, A.; Jacquet, E.; et al. The conserved regulatory RNA RsaE down-regulates the arginine degradation pathway in Staphylococcus aureus. Nucleic Acids Res. 2018, 46, 8803–8816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Treviño, J.; Ramirez-Peña, E.; Sumby, P. The small regulatory RNA FasX controls pilus expression and adherence in the human bacterial pathogen group A Streptococcus. Mol. Microbiol. 2012, 86, 140–154. [Google Scholar] [CrossRef]
- Kreikemeyer, B.; Boyle, M.D.P.; Buttaro, B.A.; Heinemann, M.; Podbielski, A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 2001, 39, 392–406. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Peña, E.; Treviño, J.; Liu, Z.; Perez, N.; Sumby, P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol. Microbiol. 2010, 78, 1332–1347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laux, A.; Sexauer, A.; Sivaselvarajah, D.; Kaysen, A.; Brückner, R. Control of competence by related non-coding csRNAs in Streptococcus pneumoniae R6. Front. Genet. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, S.A.; Scott, J.R. RivR and the small RNA RivX: The missing links between the CovR regulatory cascade and the Mga regulon. Mol. Microbiol. 2007, 66, 1506–1522. [Google Scholar] [CrossRef]
- Cheung, J.K.; Awad, M.M.; McGowan, S.; Rood, J.I. Functional Analysis of the VirSR Phosphorelay from Clostridium perfringens. PLoS ONE 2009, 4, e5849. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Yaguchi, H.; Ohtani, K.; Banu, S.; Hayashi, H. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol. Microbiol. 2002, 43, 257–265. [Google Scholar] [CrossRef]
- Ohtani, K. Gene regulation by the VirS/VirR system in Clostridium perfringens. Anaerobe 2016, 41, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Obana, N.; Shirahama, Y.; Abe, K.; Nakamura, K. Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5’ leader sequence. Mol. Microbiol. 2010, 77, 1416–1428. [Google Scholar] [CrossRef]
- Banu, S.; Ohtani, K.; Yaguchi, H.; Swe, T.; Cole, S.T.; Hayashi, H.; Shimizu, T. Identification of novel VirR/VirS-regulated genes in Clostridium perfringens. Mol. Microbiol. 2000, 35, 854–864. [Google Scholar] [CrossRef] [Green Version]
- Fang, F.C.; Frawley, E.R.; Tapscott, T.; Vazquez-Torres, A. Bacterial Stress Responses during Host Infection. Cell Host Microbe 2016, 20, 133–143. [Google Scholar] [CrossRef] [Green Version]
- Kazmierczak, M.J.; Wiedmann, M.; Boor, K.J. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. MMBR 2005, 69, 527–543. [Google Scholar] [CrossRef] [Green Version]
- Guldimann, C.; Boor, K.J.; Wiedmann, M.; Guariglia-Oropeza, V. Resilience in the Face of Uncertainty: Sigma Factor B Fine-Tunes Gene Expression to Support Homeostasis in Gram-Positive Bacteria. Appl. Environ. Microbiol. 2016, 82, 4456–4469. [Google Scholar] [CrossRef] [Green Version]
- Geissmann, T.; Chevalier, C.; Cros, M.-J.; Boisset, S.; Fechter, P.; Noirot, C.; Schrenzel, J.; François, P.; Vandenesch, F.; Gaspin, C.; et al. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009, 37, 7239–7257. [Google Scholar] [CrossRef] [PubMed]
- Romilly, C.; Lays, C.; Tomasini, A.; Caldelari, I.; Benito, Y.; Hammann, P.; Geissmann, T.; Boisset, S.; Romby, P.; Vandenesch, F. A non-coding RNA promotes bacterial persistence and decreases virulence by regulating a regulator in Staphylococcus aureus. PLoS Pathog. 2014, 10, e1003979. [Google Scholar] [CrossRef] [PubMed]
- Tomasini, A.; Moreau, K.; Chicher, J.; Geissmann, T.; Vandenesch, F.; Romby, P.; Marzi, S.; Caldelari, I. The RNA targetome of Staphylococcus aureus non-coding RNA RsaA: Impact on cell surface properties and defense mechanisms. Nucleic Acids Res. 2017, 45, 6746–6760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, J.S.; Olsen, A.S.; Bonde, M.; Valentin-Hansen, P.; Kallipolitis, B.H. Identification of a sigma B-dependent small noncoding RNA in Listeria monocytogenes. J. Bacteriol. 2008, 190, 6264–6270. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Orsi, R.H.; Boor, K.J.; Wiedmann, M.; Guariglia-Oropeza, V. Home Alone: Elimination of All but One Alternative Sigma Factor in Listeria monocytogenes Allows Prediction of New Roles for σB. Front. Microbiol. 2017, 8, 1910. [Google Scholar] [CrossRef] [Green Version]
- Mraheil, M.A.; Billion, A.; Mohamed, W.; Mukherjee, K.; Kuenne, C.; Pischimarov, J.; Krawitz, C.; Retey, J.; Hartsch, T.; Chakraborty, T.; et al. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. 2011, 39, 4235–4248. [Google Scholar] [CrossRef] [Green Version]
- Toledo-Arana, A.; Dussurget, O.; Nikitas, G.; Sesto, N.; Guet-Revillet, H.; Balestrino, D.; Loh, E.; Gripenland, J.; Tiensuu, T.; Vaitkevicius, K.; et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature 2009, 459, 950–956. [Google Scholar] [CrossRef]
- Gottesman, S. Trouble is coming: Signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem. 2019, 294, 11685–11700. [Google Scholar] [CrossRef] [Green Version]
- Rouvière, P.E.; De Las Peñas, A.; Mecsas, J.; Lu, C.Z.; Rudd, K.E.; Gross, C.A. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 1995, 14, 1032–1042. [Google Scholar] [CrossRef]
- Brooks, B.E.; Buchanan, S.K. Signaling mechanisms for activation of extracytoplasmic function (ECF) sigma factors. Biochim. Biophys. Acta 2008, 1778, 1930–1945. [Google Scholar] [CrossRef] [Green Version]
- Mutalik, V.K.; Nonaka, G.; Ades, S.E.; Rhodius, V.A.; Gross, C.A. Promoter Strength Properties of the Complete Sigma E Regulon of Escherichia coli and Salmonella enterica. J. Bacteriol. 2009, 191, 7279–7287. [Google Scholar] [CrossRef] [Green Version]
- Gogol, E.B.; Rhodius, V.A.; Papenfort, K.; Vogel, J.; Gross, C.A. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl. Acad. Sci. USA 2011, 108, 12875–12880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, J.; Rasmussen, A.A.; Overgaard, M.; Valentin-Hansen, P. Conserved small non-coding RNAs that belong to the sigmaE regulon: Role in down-regulation of outer membrane proteins. J. Mol. Biol. 2006, 364, 1–8. [Google Scholar] [CrossRef] [PubMed]
- De Las Peñas, A.; Connolly, L.; Gross, C.A. SigmaE is an essential sigma factor in Escherichia coli. J. Bacteriol. 1997, 179, 6862–6864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, M.S.; Updegrove, T.B.; Gogol, E.B.; Shabalina, S.A.; Gross, C.A.; Storz, G. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014, 28, 1620–1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, T.; Mika, F.; Lindmark, B.; Liu, Z.; Schild, S.; Bishop, A.; Zhu, J.; Camilli, A.; Johansson, J.; Vogel, J.; et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 2008, 70, 100–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmadi Badi, S.; Bruno, S.P.; Moshiri, A.; Tarashi, S.; Siadat, S.D.; Masotti, A. Small RNAs in Outer Membrane Vesicles and Their Function in Host-Microbe Interactions. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Sabharwal, D.; Wai, S.N. VrrA mediates Hfq-dependent regulation of OmpT synthesis in Vibrio cholerae. J. Mol. Biol. 2010, 400, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Sabharwal, D.; Gurung, J.M.; Cheng, A.T.; Sjöström, A.E.; Yildiz, F.H.; Uhlin, B.E.; Wai, S.N. Vibrio cholerae utilizes direct sRNA regulation in expression of a biofilm matrix protein. PLoS ONE 2014, 9, e101280. [Google Scholar] [CrossRef] [Green Version]
- Sabharwal, D.; Song, T.; Papenfort, K.; Wai, S.N. The VrrA sRNA controls a stationary phase survival factor Vrp of Vibrio cholerae. RNA Biol. 2015, 12, 186–196. [Google Scholar] [CrossRef] [Green Version]
- Peschek, N.; Hoyos, M.; Herzog, R.; Förstner, K.U.; Papenfort, K. A conserved RNA seed-pairing domain directs small RNA-mediated stress resistance in enterobacteria. EMBO J. 2019, 38, e101650. [Google Scholar] [CrossRef] [PubMed]
- Dalebroux, Z.D.; Miller, S.I. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr. Opin. Microbiol. 2014, 17, 106–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Kim, J.; Darwin, A.J. Regulation of bacterial virulence gene expression by cell envelope stress responses. Virulence 2015, 5, 835–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trimble, M.J.; Mlynárčik, P.; Kolář, M.; Hancock, R.E.W. Polymyxin: Alternative Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peschel, A. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 2002, 10, 179–186. [Google Scholar] [CrossRef]
- Alix, E.; Blanc-Potard, A.-B. Peptide-assisted degradation of the Salmonella MgtC virulence factor. EMBO J. 2008, 27, 546–557. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, W.; Zheng, K.; Liu, Z.-F. Small Non-Coding RNAs: New Insights in Modulation of Host Immune Response by Intracellular Bacterial Pathogens. Front. Immunol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Nikaido, H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [Green Version]
- Nikaido, H.; Rosenberg, E.Y. Porin channels in Escherichia coli: Studies with liposomes reconstituted from purified proteins. J. Bacteriol. 1983, 153, 241–252. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.; Bak, G.; Lee, J.; Kim, K.-S. Systematic analysis of the role of bacterial Hfq-interacting sRNAs in the response to antibiotics. J. Antimicrob. Chemother. 2015, 70, 1659–1668. [Google Scholar] [CrossRef] [Green Version]
- Felden, B.; Cattoir, V. Bacterial Adaptation to Antibiotics through Regulatory RNAs. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]
- Soutourina, O.A.; Bertin, P.N. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol. Rev. 2003, 27, 505–523. [Google Scholar] [CrossRef] [Green Version]
- Romilly, C.; Hoekzema, M.; Holmqvist, E.; Wagner, E.G.H. Small RNAs OmrA and OmrB promote class III flagellar gene expression by inhibiting the synthesis of anti-Sigma factor FlgM. RNA Biol. 2020, 17, 872–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Puyvelde, S.; Steenackers, H.P.; Vanderleyden, J. Small RNAs regulating biofilm formation and outer membrane homeostasis. RNA Biol. 2013, 10, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Svenningsen, S.L. Small RNA-Based Regulation of Bacterial Quorum Sensing and Biofilm Formation. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef]
- Benito, Y.; Kolb, F.A.; Romby, P.; Lina, G.; Etienne, J.; Vandenesch, F. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA 2000, 6, 668–679. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wu, N.; Dong, J.; Gao, Y.; Zhang, X.; Mu, C.; Shao, N.; Yang, G. Hfq Is a Global Regulator That Controls the Pathogenicity of Staphylococcus aureus. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Felden, B.; Vandenesch, F.; Bouloc, P.; Romby, P. The Staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog. 2011, 7, e1002006. [Google Scholar] [CrossRef]
- Guillet, J.; Hallier, M.; Felden, B. Emerging Functions for the Staphylococcus aureus RNome. PLoS Pathog. 2013, 9, e1003767. [Google Scholar] [CrossRef]
- Morfeldt, E.; Taylor, D.; von Gabain, A.; Arvidson, S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995, 14, 4569–4577. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Mu, C.; Ying, X.; Li, W.; Wu, N.; Dong, J.; Gao, Y.; Shao, N.; Fan, M.; Yang, G. RNAIII activates map expression by forming an RNA-RNA complex in Staphylococcus aureus. FEBS Lett. 2011, 585, 899–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boisset, S.; Geissmann, T.; Huntzinger, E.; Fechter, P.; Bendridi, N.; Possedko, M.; Chevalier, C.; Helfer, A.C.; Benito, Y.; Jacquier, A.; et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007, 21, 1353–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traber, K.E.; Lee, E.; Benson, S.; Corrigan, R.; Cantera, M.; Shopsin, B.; Novick, R.P. agr function in clinical Staphylococcus aureus isolates. Microbiology 2008, 154, 2265–2274. [Google Scholar] [CrossRef] [Green Version]
- Huntzinger, E.; Boisset, S.; Saveanu, C.; Benito, Y.; Geissmann, T.; Namane, A.; Lina, G.; Etienne, J.; Ehresmann, B.; Ehresmann, C.; et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005, 24, 824–835. [Google Scholar] [CrossRef] [Green Version]
- Chabelskaya, S.; Bordeau, V.; Felden, B. Dual RNA regulatory control of a Staphylococcus aureus virulence factor. Nucleic Acids Res. 2014, 42, 4847–4858. [Google Scholar] [CrossRef]
- Shopsin, B.; Drlica-Wagner, A.; Mathema, B.; Adhikari, R.P.; Kreiswirth, B.N.; Novick, R.P. Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J. Infect. Dis. 2008, 198, 1171–1174. [Google Scholar] [CrossRef] [Green Version]
- Shopsin, B.; Eaton, C.; Wasserman, G.A.; Mathema, B.; Adhikari, R.P.; Agolory, S.; Altman, D.R.; Holzman, R.S.; Kreiswirth, B.N.; Novick, R.P. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 2010, 202, 1593–1599. [Google Scholar] [CrossRef] [Green Version]
- Queck, S.Y.; Khan, B.A.; Wang, R.; Bach, T.-H.L.; Kretschmer, D.; Chen, L.; Kreiswirth, B.N.; Peschel, A.; Deleo, F.R.; Otto, M. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog. 2009, 5, e1000533. [Google Scholar] [CrossRef]
- Kaito, C.; Saito, Y.; Ikuo, M.; Omae, Y.; Mao, H.; Nagano, G.; Fujiyuki, T.; Numata, S.; Han, X.; Obata, K.; et al. Mobile Genetic Element SCCmec-encoded psm-mec RNA Suppresses Translation of agrA and Attenuates MRSA Virulence. PLoS Pathog. 2013, 9, e1003269. [Google Scholar] [CrossRef]
- Autret, N.; Raynaud, C.; Dubail, I.; Berche, P.; Charbit, A. Identification of the agr Locus of Listeria monocytogenes: Role in Bacterial Virulence. Infect. Immun. 2003, 71, 4463–4471. [Google Scholar] [CrossRef] [Green Version]
- Martin, M.J.; Clare, S.; Goulding, D.; Faulds-Pain, A.; Barquist, L.; Browne, H.P.; Pettit, L.; Dougan, G.; Lawley, T.D.; Wren, B.W. The agr locus regulates virulence and colonization genes in Clostridium difficile 027. J. Bacteriol. 2013, 195, 3672–3681. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Singh, K.V.; Weinstock, G.M.; Murray, B.E. Effects of Enterococcus faecalis fsr Genes on Production of Gelatinase and a Serine Protease and Virulence. Infect. Immun. 2000, 68, 2579–2586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooksley, C.M.; Davis, I.J.; Winzer, K.; Chan, W.C.; Peck, M.W.; Minton, N.P. Regulation of Neurotoxin Production and Sporulation by a Putative agrBD Signaling System in Proteolytic Clostridium botulinum. Appl. Environ. Microbiol. 2010, 76, 4448–4460. [Google Scholar] [CrossRef] [Green Version]
- Ohtani, K.; Yuan, Y.; Hassan, S.; Wang, R.; Wang, Y.; Shimizu, T. Virulence Gene Regulation by the agr System in Clostridium perfringens. J. Bacteriol. 2009, 191, 3919–3927. [Google Scholar] [CrossRef] [Green Version]
- Bernheim, A.; Sorek, R. The pan-immune system of bacteria: Antiviral defence as a community resource. Nat. Rev. Microbiol. 2020, 18, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Patterson, A.G.; Jackson, S.A.; Taylor, C.; Evans, G.B.; Salmond, G.P.C.; Przybilski, R.; Staals, R.H.J.; Fineran, P.C. Quorum Sensing Controls Adaptive Immunity through the Regulation of Multiple CRISPR-Cas Systems. Mol. Cell 2016, 64, 1102–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Høyland-Kroghsbo, N.M.; Paczkowski, J.; Mukherjee, S.; Broniewski, J.; Westra, E.; Bondy-Denomy, J.; Bassler, B.L. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc. Natl. Acad. Sci. USA 2017, 114, 131–135. [Google Scholar] [CrossRef] [Green Version]
- Mion, S.; Plener, L.; Rémy, B.; Daudé, D.; Chabrière, É. Lactonase SsoPox modulates CRISPR-Cas expression in gram-negative proteobacteria using AHL-based quorum sensing systems. Res. Microbiol. 2019, 170, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Serbanescu, M.A.; Cordova, M.; Krastel, K.; Flick, R.; Beloglazova, N.; Latos, A.; Yakunin, A.F.; Senadheera, D.B.; Cvitkovitch, D.G. Role of the Streptococcus mutans CRISPR-Cas systems in immunity and cell physiology. J. Bacteriol. 2015, 197, 749–761. [Google Scholar] [CrossRef] [Green Version]
- Marx, P.; Nuhn, M.; Kovács, M.; Hakenbeck, R.; Brückner, R. Identification of genes for small non-coding RNAs that belong to the regulon of the two-component regulatory system CiaRH in Streptococcus. BMC Genomics 2010, 11, 661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Høyland-Kroghsbo, N.M.; Muñoz, K.A.; Bassler, B.L. Temperature, by Controlling Growth Rate, Regulates CRISPR-Cas Activity in Pseudomonas aeruginosa. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westra, E.R.; Buckling, A.; Fineran, P.C. CRISPR-Cas systems: Beyond adaptive immunity. Nat. Rev. Microbiol. 2014, 12, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Weiss, D.S. CRISPR-Cas systems: New players in gene regulation and bacterial physiology. Front. Cell. Infect. Microbiol. 2014, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Saroj, S.D.; Llewellyn, A.C.; Tzeng, Y.-L.; Weiss, D.S. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 2013, 497, 254–257. [Google Scholar] [CrossRef] [Green Version]
- Sesto, N.; Touchon, M.; Andrade, J.M.; Kondo, J.; Rocha, E.P.C.; Arraiano, C.M.; Archambaud, C.; Westhof, É.; Romby, P.; Cossart, P. A PNPase dependent CRISPR System in Listeria. PLoS Genet. 2014, 10, e1004065. [Google Scholar] [CrossRef] [Green Version]
- Dugar, G.; Leenay, R.T.; Eisenbart, S.K.; Bischler, T.; Aul, B.U.; Beisel, C.L.; Sharma, C.M. CRISPR RNA-Dependent Binding and Cleavage of Endogenous RNAs by the Campylobacter jejuni Cas9. Mol. Cell. 2018, 69, 893–905.e7. [Google Scholar] [CrossRef] [Green Version]
- Cui, L.; Wang, X.; Huang, D.; Zhao, Y.; Feng, J.; Lu, Q.; Pu, Q.; Wang, Y.; Cheng, G.; Wu, M.; et al. CRISPR-cas3 of Salmonella Upregulates Bacterial Biofilm Formation and Virulence to Host Cells by Targeting Quorum-Sensing Systems. Pathogens 2020, 9, 53. [Google Scholar] [CrossRef] [Green Version]
- Brantl, S.; Brückner, R. Small regulatory RNAs from low-GC Gram-positive bacteria. RNA Biol. 2014, 11, 443–456. [Google Scholar] [CrossRef] [Green Version]
- Acebo, P.; Martin-Galiano, A.J.; Navarro, S.; Zaballos, A.; Amblar, M. Identification of 88 regulatory small RNAs in the TIGR4 strain of the human pathogen Streptococcus pneumoniae. RNA 2012, 18, 530–546. [Google Scholar] [CrossRef] [Green Version]
- Schnorpfeil, A.; Kranz, M.; Kovács, M.; Kirsch, C.; Gartmann, J.; Brunner, I.; Bittmann, S.; Brückner, R. Target evaluation of the non-coding csRNAs reveals a link of the two-component regulatory system CiaRH to competence control in Streptococcus pneumoniae R6. Mol. Microbiol. 2013, 89, 334–349. [Google Scholar] [CrossRef] [PubMed]
- Serganov, A.; Nudler, E. A decade of riboswitches. Cell 2013, 152, 17–24. [Google Scholar] [CrossRef] [Green Version]
- McCown, P.J.; Corbino, K.A.; Stav, S.; Sherlock, M.E.; Breaker, R.R. Riboswitch diversity and distribution. RNA 2017, 23, 995–1011. [Google Scholar] [CrossRef] [PubMed]
- Abduljalil, J.M. Bacterial riboswitches and RNA thermometers: Nature and contributions to pathogenesis. Non Coding RNA Res. 2018, 3, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Valentini, M.; Filloux, A. Multiple Roles of c-di-GMP Signaling in Bacterial Pathogenesis. Annu. Rev. Microbiol. 2019, 73, 387–406. [Google Scholar] [CrossRef]
- Bordeleau, E.; Fortier, L.-C.; Malouin, F.; Burrus, V. c-di-GMP turn-over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 2011, 7, e1002039. [Google Scholar] [CrossRef] [Green Version]
- Soutourina, O.A.; Monot, M.; Boudry, P.; Saujet, L.; Pichon, C.; Sismeiro, O.; Semenova, E.; Severinov, K.; Le Bouguenec, C.; Coppée, J.-Y.; et al. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 2013, 9, e1003493. [Google Scholar] [CrossRef]
- Sudarsan, N.; Lee, E.R.; Weinberg, Z.; Moy, R.H.; Kim, J.N.; Link, K.H.; Breaker, R.R. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 2008, 321, 411–413. [Google Scholar] [CrossRef] [Green Version]
- Purcell, E.B.; McKee, R.W.; McBride, S.M.; Waters, C.M.; Tamayo, R. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J. Bacteriol. 2012, 194, 3307–3316. [Google Scholar] [CrossRef] [Green Version]
- Smits, W.K.; Lyras, D.; Lacy, D.B.; Wilcox, M.H.; Kuijper, E.J. Clostridium difficile infection. Nat. Rev. Dis. Primers 2016, 2, 16020. [Google Scholar] [CrossRef] [Green Version]
- Sekulovic, O.; Mathias Garrett, E.; Bourgeois, J.; Tamayo, R.; Shen, A.; Camilli, A. Genome-wide detection of conservative site-specific recombination in bacteria. PLoS Genet. 2018, 14, e1007332. [Google Scholar] [CrossRef] [PubMed]
- Garrett, E.M.; Sekulovic, O.; Wetzel, D.; Jones, J.B.; Edwards, A.N.; Vargas-Cuebas, G.; McBride, S.M.; Tamayo, R. Phase variation of a signal transduction system controls Clostridioides difficile colony morphology, motility, and virulence. PLoS Biol. 2019, 17, e3000379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DebRoy, S.; Gebbie, M.; Ramesh, A.; Goodson, J.R.; Cruz, M.R.; van Hoof, A.; Winkler, W.C.; Garsin, D.A. Riboswitches. A riboswitch-containing sRNA controls gene expression by sequestration of a response regulator. Science 2014, 345, 937–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellin, J.R.; Koutero, M.; Dar, D.; Nahori, M.-A.; Sorek, R.; Cossart, P. Riboswitches. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science 2014, 345, 940–943. [Google Scholar] [CrossRef] [Green Version]
- Shu, C.J.; Zhulin, I.B. ANTAR: An RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci. 2002, 27, 3–5. [Google Scholar] [CrossRef]
- Danger, J.L.; Makthal, N.; Kumaraswami, M.; Sumby, P. The FasX Small Regulatory RNA Negatively Regulates the Expression of Two Fibronectin-Binding Proteins in Group A Streptococcus. J. Bacteriol. 2015, 197, 3720–3730. [Google Scholar] [CrossRef] [Green Version]
- Perez, N.; Treviño, J.; Liu, Z.; Ho, S.C.M.; Babitzke, P.; Sumby, P. A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus. PLoS ONE 2009, 4, e7668. [Google Scholar] [CrossRef] [Green Version]
- Danger, J.L.; Cao, T.N.; Cao, T.H.; Sarkar, P.; Treviño, J.; Pflughoeft, K.J.; Sumby, P. The small regulatory RNA FasX enhances group A Streptococcus virulence and inhibits pilus expression via serotype-specific targets. Mol. Microbiol. 2015, 96, 249–262. [Google Scholar] [CrossRef] [Green Version]
- Dalton, T.L.; Collins, J.T.; Barnett, T.C.; Scott, J.R. RscA, a member of the MDR1 family of transporters, is repressed by CovR and required for growth of Streptococcus pyogenes under heat stress. J. Bacteriol. 2006, 188, 77–85. [Google Scholar] [CrossRef] [Green Version]
- Roberts, S.A.; Churchward, G.G.; Scott, J.R. Unraveling the regulatory network in Streptococcus pyogenes: The global response regulator CovR represses rivR directly. J. Bacteriol. 2007, 189, 1459–1463. [Google Scholar] [CrossRef] [Green Version]
- Treviño, J.; Liu, Z.; Cao, T.N.; Ramirez-Peña, E.; Sumby, P. RivR is a negative regulator of virulence factor expression in group A Streptococcus. Infect. Immun. 2013, 81, 364–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, A.N.; de Mets, F.; Brinsmade, S.R. Who’s in control? Regulation of metabolism and pathogenesis in space and time. Curr. Opin. Microbiol. 2020, 55, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Mandin, P.; Gottesman, S. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA / DpiB two component system. Mol. Microbiol. 2009, 72, 551–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Y.; Updegrove, T.B.; Livingston, N.N.; Storz, G. Protection against deleterious nitrogen compounds: Role of σS-dependent small RNAs encoded adjacent to sdiA. Nucleic Acids Res. 2016, 44, 6935–6948. [Google Scholar] [CrossRef] [Green Version]
- Coornaert, A.; Lu, A.; Mandin, P.; Springer, M.; Gottesman, S.; Guillier, M. MicA sRNA links the PhoP regulon to cell envelope stress. Mol. Microbiol. 2010, 76, 467–479. [Google Scholar] [CrossRef] [Green Version]
- Coornaert, A.; Chiaruttini, C.; Springer, M.; Guillier, M. Post-Transcriptional Control of the Escherichia coli PhoQ-PhoP Two-Component System by Multiple sRNAs Involves a Novel Pairing Region of GcvB. PLoS Genet. 2013, 9, e1003156. [Google Scholar] [CrossRef] [Green Version]
- Shimoni, Y.; Friedlander, G.; Hetzroni, G.; Niv, G.; Altuvia, S.; Biham, O.; Margalit, H. Regulation of gene expression by small non-coding RNAs: A quantitative view. Mol. Syst. Biol. 2007, 3, 138. [Google Scholar] [CrossRef]
- Levine, E.; Hwa, T. Small RNAs establish gene expression thresholds. Curr. Opin. Microbiol. 2008, 11, 574–579. [Google Scholar] [CrossRef] [Green Version]
- Gottesman, S.; Storz, G. Bacterial Small RNA Regulators: Versatile Roles and Rapidly Evolving Variations. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [Green Version]
- Bossi, L.; Figueroa-Bossi, N. Competing endogenous RNAs: A target-centric view of small RNA regulation in bacteria. Nat. Rev. Microbiol. 2016, 14, 775–784. [Google Scholar] [CrossRef] [Green Version]
- Moon, K.; Gottesman, S. Competition among Hfq-binding small RNAs in Escherichia coli: Competition for Hfq in E. coli. Mol. Microbiol. 2011, 82, 1545–1562. [Google Scholar] [CrossRef] [PubMed]
- Sorek, R.; Cossart, P. Prokaryotic transcriptomics: A new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 2010, 11, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Hör, J.; Gorski, S.A.; Vogel, J. Bacterial RNA Biology on a Genome Scale. Mol. Cell 2018, 70, 785–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georg, J.; Lalaouna, D.; Hou, S.; Lott, S.C.; Caldelari, I.; Marzi, S.; Hess, W.R.; Romby, P. The power of cooperation: Experimental and computational approaches in the functional characterization of bacterial sRNAs. Mol. Microbiol. 2020, 113, 603–612. [Google Scholar] [CrossRef]
- Jagodnik, J.; Brosse, A.; Le Lam, T.N.; Chiaruttini, C.; Guillier, M. Mechanistic study of base-pairing small regulatory RNAs in bacteria. Methods 2017, 117, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Desgranges, E.; Caldelari, I.; Marzi, S.; Lalaouna, D. Navigation through the twists and turns of RNA sequencing technologies: Application to bacterial regulatory RNAs. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194506. [Google Scholar] [CrossRef]
- Sharma, C.M.; Hoffmann, S.; Darfeuille, F.; Reignier, J.; Findeiss, S.; Sittka, A.; Chabas, S.; Reiche, K.; Hackermüller, J.; Reinhardt, R.; et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 2010, 464, 250–255. [Google Scholar] [CrossRef]
- Wurtzel, O.; Sapra, R.; Chen, F.; Zhu, Y.; Simmons, B.A.; Sorek, R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010, 20, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Dar, D.; Sorek, R. High-resolution RNA 3’-ends mapping of bacterial Rho-dependent transcripts. Nucleic Acids Res. 2018, 46, 6797–6805. [Google Scholar] [CrossRef] [Green Version]
- Wurtzel, O.; Sesto, N.; Mellin, J.R.; Karunker, I.; Edelheit, S.; Bécavin, C.; Archambaud, C.; Cossart, P.; Sorek, R. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol. 2012, 8, 583. [Google Scholar] [CrossRef]
- Dar, D.; Shamir, M.; Mellin, J.R.; Koutero, M.; Stern-Ginossar, N.; Cossart, P.; Sorek, R. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 2016, 352, aad9822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wurtzel, O.; Yoder-Himes, D.R.; Han, K.; Dandekar, A.A.; Edelheit, S.; Greenberg, E.P.; Sorek, R.; Lory, S. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog. 2012, 8, e1002945. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Lozano, M.; Marvig, R.L.; Molin, S.; Long, K.S. Genome-wide identification of novel small RNAs in Pseudomonas aeruginosa. Environ. Microbiol. 2012, 14, 2006–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahr, T.; Rusniok, C.; Dervins-Ravault, D.; Sismeiro, O.; Coppee, J.-Y.; Buchrieser, C. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol. 2012, 9, 503–519. [Google Scholar] [CrossRef] [Green Version]
- Wilms, I.; Overlöper, A.; Nowrousian, M.; Sharma, C.M.; Narberhaus, F. Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens. RNA Biol. 2012, 9, 446–457. [Google Scholar] [CrossRef] [Green Version]
- Miotto, P.; Forti, F.; Ambrosi, A.; Pellin, D.; Veiga, D.F.; Balazsi, G.; Gennaro, M.L.; Di Serio, C.; Ghisotti, D.; Cirillo, D.M. Genome-wide discovery of small RNAs in Mycobacterium tuberculosis. PLoS ONE 2012, 7, e51950. [Google Scholar] [CrossRef] [Green Version]
- Dugar, G.; Herbig, A.; Förstner, K.U.; Heidrich, N.; Reinhardt, R.; Nieselt, K.; Sharma, C.M. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 2013, 9, e1003495. [Google Scholar] [CrossRef]
- Dequivre, M.; Diel, B.; Villard, C.; Sismeiro, O.; Durot, M.; Coppée, J.Y.; Nesme, X.; Vial, L.; Hommais, F. Small RNA Deep-Sequencing Analyses Reveal a New Regulator of Virulence in Agrobacterium fabrum C58. Mol. Plant Microbe Interact. 2015, 28, 580–589. [Google Scholar] [CrossRef] [Green Version]
- Rosinski-Chupin, I.; Sauvage, E.; Sismeiro, O.; Villain, A.; Da Cunha, V.; Caliot, M.-E.; Dillies, M.-A.; Trieu-Cuot, P.; Bouloc, P.; Lartigue, M.-F.; et al. Single nucleotide resolution RNA-seq uncovers new regulatory mechanisms in the opportunistic pathogen Streptococcus agalactiae. BMC Genomics 2015, 16, 419. [Google Scholar] [CrossRef] [Green Version]
- Papenfort, K.; Förstner, K.U.; Cong, J.-P.; Sharma, C.M.; Bassler, B.L. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc. Natl. Acad. Sci. USA 2015, 112, E766–E775. [Google Scholar] [CrossRef] [Green Version]
- Carroll, R.K.; Weiss, A.; Broach, W.H.; Wiemels, R.E.; Mogen, A.B.; Rice, K.C.; Shaw, L.N. Genome-wide Annotation, Identification, and Global Transcriptomic Analysis of Regulatory or Small RNA Gene Expression in Staphylococcus aureus. mBio 2016, 7, e01990-01915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidrich, N.; Bauriedl, S.; Barquist, L.; Li, L.; Schoen, C.; Vogel, J. The primary transcriptome of Neisseria meningitidis and its interaction with the RNA chaperone Hfq. Nucleic Acids Res. 2017, 45, 6147–6167. [Google Scholar] [CrossRef] [PubMed]
- Zhukova, A.; Fernandes, L.G.; Hugon, P.; Pappas, C.J.; Sismeiro, O.; Coppée, J.-Y.; Becavin, C.; Malabat, C.; Eshghi, A.; Zhang, J.-J.; et al. Genome-Wide Transcriptional Start Site Mapping and sRNA Identification in the Pathogen Leptospira interrogans. Front. Cell. Infect. Microbiol. 2017, 7, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kröger, C.; MacKenzie, K.D.; Alshabib, E.Y.; Kirzinger, M.W.B.; Suchan, D.M.; Chao, T.-C.; Akulova, V.; Miranda-CasoLuengo, A.A.; Monzon, V.A.; Conway, T.; et al. The primary transcriptome, small RNAs and regulation of antimicrobial resistance in Acinetobacter baumannii ATCC 17978. Nucleic Acids Res. 2018, 46, 9684–9698. [Google Scholar] [CrossRef] [Green Version]
- Rosinski-Chupin, I.; Sauvage, E.; Fouet, A.; Poyart, C.; Glaser, P. Conserved and specific features of Streptococcus pyogenes and Streptococcus agalactiae transcriptional landscapes. BMC Genomics 2019, 20, 236. [Google Scholar] [CrossRef] [PubMed]
- Sinha, D.; Zimmer, K.; Cameron, T.A.; Rusch, D.B.; Winkler, M.E.; De Lay, N.R. Redefining the Small Regulatory RNA Transcriptome in Streptococcus pneumoniae Serotype 2 Strain D39. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [Green Version]
- Lécrivain, A.-L.; Beckmann, B.M. Bacterial RNA in extracellular vesicles: A new regulator of host-pathogen interactions? Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194519. [Google Scholar] [CrossRef]
- Koeppen, K.; Hampton, T.H.; Jarek, M.; Scharfe, M.; Gerber, S.A.; Mielcarz, D.W.; Demers, E.G.; Dolben, E.L.; Hammond, J.H.; Hogan, D.A.; et al. A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles. PLoS Pathog. 2016, 12, e1005672. [Google Scholar] [CrossRef]
- Riedel, C.U.; Monk, I.R.; Casey, P.G.; Waidmann, M.S.; Gahan, C.G.M.; Hill, C. AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in Listeria monocytogenes. Mol. Microbiol. 2009, 71, 1177–1189. [Google Scholar] [CrossRef]
- Woodson, S.A.; Panja, S.; Santiago-Frangos, A. Proteins That Chaperone RNA Regulation. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Trinquier, A.; Durand, S.; Braun, F.; Condon, C. Regulation of RNA processing and degradation in bacteria. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194505. [Google Scholar] [CrossRef] [PubMed]
- Romeo, T.; Babitzke, P. Global Regulation by CsrA and Its RNA Antagonists. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [Green Version]
- Pichon, C.; du Merle, L.; Caliot, M.E.; Trieu-Cuot, P.; Le Bouguénec, C. An in silico model for identification of small RNAs in whole bacterial genomes: Characterization of antisense RNAs in pathogenic Escherichia coli and Streptococcus agalactiae strains. Nucleic Acids Res. 2012, 40, 2846–2861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pain, A.; Ott, A.; Amine, H.; Rochat, T.; Bouloc, P.; Gautheret, D. An assessment of bacterial small RNA target prediction programs. RNA Biol. 2015, 12, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.R.; Mann, M.; Backofen, R. Structure and Interaction Prediction in Prokaryotic RNA Biology. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [Green Version]
- Lalaouna, D.; Carrier, M.-C.; Semsey, S.; Brouard, J.-S.; Wang, J.; Wade, J.T.; Massé, E. A 3’ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol. Cell. 2015, 58, 393–405. [Google Scholar] [CrossRef] [Green Version]
- Lalaouna, D.; Prévost, K.; Eyraud, A.; Massé, E. Identification of unknown RNA partners using MAPS. Methods 2017, 117, 28–34. [Google Scholar] [CrossRef]
- Melamed, S.; Faigenbaum-Romm, R.; Peer, A.; Reiss, N.; Shechter, O.; Bar, A.; Altuvia, Y.; Argaman, L.; Margalit, H. Mapping the small RNA interactome in bacteria using RIL-seq. Nat. Protoc. 2018, 13, 1–33. [Google Scholar] [CrossRef]
- Waters, S.A.; McAteer, S.P.; Kudla, G.; Pang, I.; Deshpande, N.P.; Amos, T.G.; Leong, K.W.; Wilkins, M.R.; Strugnell, R.; Gally, D.L.; et al. Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E. EMBO J. 2017, 36, 374–387. [Google Scholar] [CrossRef]
- Okumura, K.; Ohtani, K.; Hayashi, H.; Shimizu, T. Characterization of genes regulated directly by the VirR/VirS system in Clostridium perfringens. J. Bacteriol. 2008, 190, 7719–7727. [Google Scholar] [CrossRef] [Green Version]
- Wade, J.T. Mapping Transcription Regulatory Networks with ChIP-seq and RNA-seq. Adv. Exp. Med. Biol. 2015, 883, 119–134. [Google Scholar] [CrossRef]
- Mann, B.; van Opijnen, T.; Wang, J.; Obert, C.; Wang, Y.-D.; Carter, R.; McGoldrick, D.J.; Ridout, G.; Camilli, A.; Tuomanen, E.I.; et al. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog. 2012, 8, e1002788. [Google Scholar] [CrossRef]
- Klemm, E.J.; Wong, V.K.; Dougan, G. Emergence of dominant multidrug-resistant bacterial clades: Lessons from history and whole-genome sequencing. Proc. Natl. Acad. Sci. USA 2018, 115, 12872–12877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peacock, S.J.; Paterson, G.K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Lynch, S.V. The gut microbiome: Relationships with disease and opportunities for therapy. J. Exp. Med. 2019, 216, 20–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- McInnes, R.S.; McCallum, G.E.; Lamberte, L.E.; van Schaik, W. Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr. Opin. Microbiol. 2020, 53, 35–43. [Google Scholar] [CrossRef]
- Tiwari, S.; Jamal, S.B.; Hassan, S.S.; Carvalho, P.V.S.D.; Almeida, S.; Barh, D.; Ghosh, P.; Silva, A.; Castro, T.L.P.; Azevedo, V. Two-Component Signal Transduction Systems of Pathogenic Bacteria As Targets for Antimicrobial Therapy: An Overview. Front. Microbiol. 2017, 8, 1878. [Google Scholar] [CrossRef]
- Bem, A.E.; Velikova, N.; Pellicer, M.T.; van Baarlen, P.; Marina, A.; Wells, J.M. Bacterial Histidine Kinases as Novel Antibacterial Drug Targets. ACS Chem. Biol. 2015, 10, 213–224. [Google Scholar] [CrossRef]
- Rasko, D.A.; Moreira, C.G.; Li, D.R.; Reading, N.C.; Ritchie, J.M.; Waldor, M.K.; Williams, N.; Taussig, R.; Wei, S.; Roth, M.; et al. Targeting QseC Signaling and Virulence for Antibiotic Development. Science 2008, 321, 1078–1080. [Google Scholar] [CrossRef] [Green Version]
- Barra, M.; Danino, T.; Garrido, D. Engineered Probiotics for Detection and Treatment of Inflammatory Intestinal Diseases. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Chen, X.; Sheng, H.; Shen, X.; Sun, X.; Yan, Y.; Wang, J.; Yuan, Q. Engineering probiotics as living diagnostics and therapeutics for improving human health. Microb. Cell Fact. 2020, 19, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, J.D.; Piattelli, E.; McCormick, B.A.; Silby, M.W.; Brigham, C.J.; Bucci, V. Engineered Probiotic for the Inhibition of Salmonella via Tetrathionate-Induced Production of Microcin H47. ACS Infect. Dis. 2018, 4, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Witztum, J.L.; Bennett, C.F.; Baker, B.F. RNA-Targeted Therapeutics. Cell Metab. 2018, 27, 714–739. [Google Scholar] [CrossRef] [PubMed]
- Sully, E.K.; Geller, B.L. Antisense antimicrobial therapeutics. Curr. Opin. Microbiol. 2016, 33, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Sawyer, A.J.; Wesolowski, D.; Gandotra, N.; Stojadinovic, A.; Izadjoo, M.; Altman, S.; Kyriakides, T.R. A peptide-morpholino oligomer conjugate targeting Staphylococcus aureus gyrA mRNA improves healing in an infected mouse cutaneous wound model. Int. J. Pharm. 2013, 453, 651–655. [Google Scholar] [CrossRef] [Green Version]
- Law, C.O.K.; Huang, C.; Pan, Q.; Lee, J.; Hao, Q.; Chan, T.-F.; Lo, N.W.S.; Ang, I.L.; Koon, A.; Ip, M.; et al. A Small RNA Transforms the Multidrug Resistance of Pseudomonas aeruginosa to Drug Susceptibility. Mol. Ther. Nucleic Acids 2019, 16, 218–228. [Google Scholar] [CrossRef] [Green Version]
- Xue, X.-Y.; Mao, X.-G.; Zhou, Y.; Chen, Z.; Hu, Y.; Hou, Z.; Li, M.-K.; Meng, J.-R.; Luo, X.-X. Advances in the delivery of antisense oligonucleotides for combating bacterial infectious diseases. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 745–758. [Google Scholar] [CrossRef]
- Vogel, J. An RNA biology perspective on species-specific programmable RNA antibiotics. Mol. Microbiol. 2020, 113, 550–559. [Google Scholar] [CrossRef] [Green Version]
- Dersch, P.; Khan, M.A.; Mühlen, S.; Görke, B. Roles of Regulatory RNAs for Antibiotic Resistance in Bacteria and Their Potential Value as Novel Drug Targets. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petchiappan, A.; Chatterji, D. Antibiotic Resistance: Current Perspectives. ACS Omega 2017, 2, 7400–7409. [Google Scholar] [CrossRef] [PubMed]
- El-Mowafi, S.A.; Alumasa, J.N.; Ades, S.E.; Keiler, K.C. Cell-based assay to identify inhibitors of the Hfq-sRNA regulatory pathway. Antimicrob. Agents Chemother. 2014, 58, 5500–5509. [Google Scholar] [CrossRef] [Green Version]
- Bikard, D.; Barrangou, R. Using CRISPR-Cas systems as antimicrobials. Curr. Opin. Microbiol. 2017, 37, 155–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mion, S.; Rémy, B.; Plener, L.; Brégeon, F.; Chabrière, E.; Daudé, D. Quorum Quenching Lactonase Strengthens Bacteriophage and Antibiotic Arsenal Against Pseudomonas aeruginosa Clinical Isolates. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Blount, K.F.; Breaker, R.R. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006, 24, 1558–1564. [Google Scholar] [CrossRef]
- Lünse, C.E.; Schüller, A.; Mayer, G. The promise of riboswitches as potential antibacterial drug targets. Int. J. Med. Microbiol. 2014, 304, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Peltier, J.; Soutourina, O. Identification of c-di-GMP-Responsive Riboswitches. In c-di-GMP Signaling; Sauer, K., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 377–402. ISBN 978-1-4939-7239-5. [Google Scholar]
- McKee, R.W.; Harvest, C.K.; Tamayo, R. Cyclic Diguanylate Regulates Virulence Factor Genes via Multiple Riboswitches in Clostridium difficile. mSphere 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.-H.; Le Roux, A.; Boyapelly, K.; Lamontagne, A.-M.; Archambault, M.-A.; Picard-Jean, F.; Lalonde-Seguin, D.; St-Pierre, E.; Najmanovich, R.J.; Fortier, L.-C.; et al. Purine analogs targeting the guanine riboswitch as potential antibiotics against Clostridioides difficile. Eur. J. Med. Chem. 2018, 143, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Mulhbacher, J.; Brouillette, E.; Allard, M.; Fortier, L.-C.; Malouin, F.; Lafontaine, D.A. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 2010, 6, e1000865. [Google Scholar] [CrossRef] [Green Version]
- Ster, C.; Allard, M.; Boulanger, S.; Lamontagne Boulet, M.; Mulhbacher, J.; Lafontaine, D.A.; Marsault, E.; Lacasse, P.; Malouin, F. Experimental treatment of Staphylococcus aureus bovine intramammary infection using a guanine riboswitch ligand analog. J. Dairy Sci. 2013, 96, 1000–1008. [Google Scholar] [CrossRef] [Green Version]
- Blount, K.F.; Megyola, C.; Plummer, M.; Osterman, D.; O’Connell, T.; Aristoff, P.; Quinn, C.; Chrusciel, R.A.; Poel, T.J.; Schostarez, H.J.; et al. Novel riboswitch-binding flavin analog that protects mice against Clostridium difficile infection without inhibiting cecal flora. Antimicrob. Agents Chemother. 2015, 59, 5736–5746. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Pathogen | TCS | sRNA Regulated by TCS a | Targets b | Roles in Virulence | Stimuli | References | |
|---|---|---|---|---|---|---|---|
| HK | RR | ||||||
| Gram-negative bacteria | |||||||
| Escherichia coli, Salmonella enterica serovar Typhimurium | CpxA | CpxR | (+) CpxQ (−) CyaR (+) RprA | CpxQ: nhaB (Na-H antiporter), skp (chaperone protein), agp (periplasmic acid glucose-1-phosphatase), fimA (major fimbrin subunit of type 1 pilus) and ydjN (cystine transporter); CyaR: ompX (outer membrane protein), luxS (autoinducer-2 synthase), nadE (essential NAD synthetase) and yqaE (membrane protein with unknown function), hdeD (acid-resistance membrane protein); RprA (see below); | Envelope-stress response | High pH, osmolality, alteration in IM lipid composition | [25,26,27,28,29,30] |
| EnvZ | OmpR | (+) OmrA (+) OmrB (+) MicF | OmrA/OmrB: ompT (outer membrane protease), cirA, fecA, fepA, (receptors for iron–siderophore complexes), csgD (TR of curli genes), flhDC (master regulator of flagellar synthesis) and ompR-envZ (TCS); MicF: ompF (outer membrane protein), cpxA-cpxR (TCS), lrp (TR); | Envelope-stress response | Osmolality | [31,32,33,34,35,36,37] | |
| PhoQ | PhoP | (+) MgrR, (+) AmgR (in Salmonella) (+) PinT (in Salmonella) | MgrR: eptB (phosphoethanolamine transferase), ygdQ (hypothetical protein) and soxS (TR involved in oxidative stress); AmgR: mgtC (virulence protein); PinT: hilA (hyperinvasion locus A) and rtsA (TR) | Resistance to CAMPs, survival within macrophages, expression of type 3 secretion system (T3SS) | Low Mg2+/Ca2+, antimicrobial peptides | [38,39,40,41,42,43] | |
| RcsC RcsD c | RcsB | (+) RprA | rpoS (central regulator of the general stress response), csgD (TR of curli genes) in S. Typhimurium, hdeD (acid-resistance membrane protein); | Inhibition of biofilm development and stress response | Alterations of bacterial envelope | [30,44,45,46] | |
| GlrK (QseE YfhK) | GlrR (QseF, YfhA) | (+) GlmY and GlmZ | GlmY: GlmZ; GlmZ: glmS (glucosamine-6-phosphate synthase), Locus of Enterocyte Effacement (LEE) 4 and LEE5 operons, espFu and nleA (non-LEE-encoded effectors) | Antibiotic resistance, attaching and effacing (AE) lesion formations | Host hormones such as epinephrine and norepinephrine in Enterohemorrhagic E. coli (EHEC) | [47,48,49,50] | |
| QseC | QseB | (+) GlmY | Autoinducer 3 (AI-3) and the adrenergic hormones | [51] | |||
| ArcB | ArcA | (−) ArcZ | rpoS (central regulator of the general stress response), agrB-agrA (TCS), flhDC (master regulator of flagellar genes); | Regulation of motility, host adaptation | Aerobic conditions | [34,52] | |
| Helicobacter pylori | ArsS | ArsR | Cis-antisense 5′ureB-sRNA | ureAB (urease) | Colonization of the gastric mucosa | Low pH | [53] |
| Vibrio cholerae | LuxPQ d | Lux O | (+) Qrr1-4 | hapR (master TR), large type 6 secretion system cluster | Expression of virulence and biofilm genes | Cell density—AI2 | [54,55,56,57] |
| CqsS | LuxO | ||||||
| EnvZ | OmpR | (+) CoaR | tcpI (TR) | Expression of major pilin subunit | Osmolality | [58,59] | |
| Pseudomonas aeruginosa | CbrA | CbrB | (+)CrcZ | Sequestration of the RNA-binding protein Crc | Metabolism, susceptibility to antibiotics and virulence | Carbon and nitrogen source | [60,61] |
| NtrB | NtrC | (+)NrsZ | rhlA (rhamnolipid biosynthesis) | Increases production of the virulence factor rhamnolipid | Nitrogen limitation | [62] | |
| NarX | NarL | (+)PaiI | Unknown | Colonization of tumor | Low oxygen and nitrate | [63] | |
| KinB | AlgB | Type I CRISPR-Cas | Immunity to phage infection | Unknown | [64] | ||
| PmrA | PmrB | (+) RsmY | Sequestration of the RNA-binding protein RsmA | Indirect effect on the expression of lipA (lipase) | Low Mg 2+ | [65] | |
| TCS BarA-SirA and its homologous systems | |||||||
| S. typhimurium | BarA | SirA | (+) CsrB (+) CsrC | Sequestration of the RNA-binding protein CsrA/RsmA | Metabolism, motility, biofilm formation, stress resistance, virulence and quorum sensing | BarA senses the presence of carboxylate compounds. | [54,66,67,68,69,70,71] |
| E. coli | BarA | UvrY | |||||
| P. aeruginosa | GacS | GacA | (+) RsmY (+) RsmZ | ||||
| Legionella pneumophila | LetS | LetA | (+) RsmY (+) RsmZ | ||||
| V. cholerae | VarS | VarA | (+) CsrB (+) CsrC (+) CsrD | ||||
| Gram-Positive Bacteria | |||||||
| Listeria monocytogenes | LisK | LisR | (+) LhrC 1–5 | lapB (virulence adhesin), tcsA (T cell-stimulating antigen), oppA (oligo-peptide binding protein) | Adhesion and invasion of non-phagocytic cells, expression of T cell-stimulating antigen TcsA | Cell envelope stress | [72,73,74,75,76] |
| LisK | LisR | (+) Rli22 | oppA (oligo-peptide binding protein) | Expression of virulence-associated oligo-peptide binding protein | Cell envelope stress | [77] | |
| VirS | VirR | (+) Rli32 | Inducer of IFN-β expression; lhrC locus (LhrC1-5) | Induction of IFN-β expression | In vivo infection | [78,79] | |
| Mycobacterium tuberculosis | PhoR | PhoP | (+) Mcr7 | tatC (Twin Arginine Translocation (Tat) protein secretion apparatus) | Secretion of the immunodominant Ag85 complex and the beta-lactamase BlaC | Unknown | [80] |
| Staphylococcus aureus | AgrC | AgrA | (+) RNAIII (+) ArtR | RNAIII: rot, mgrA (master TRs); spa, coa, sbi, Sa1000, lytM, hla (virulence factors); ArtR: sarT (TR) | Evasion of host immunity, toxins expression | Cell density (autoinducers molecule) | [81,82] |
| α-toxin expression | Cell density | [83] | |||||
| SsrB | SsrA | (+) RsaE | opp3A (ABC transporter component) rocF (arginase involved in the arginine catabolism). In Staphylococcus epidermidis. lrgA (antiholin) icaR (icaADBC biofilm operon repressor) and sucCD (succinyl-CoA synthetase) | Metabolic adaptation, biofilm formation, eDNA release. | Low O2 and NO | [84,85,86,87] | |
| Group A Streptococcus | FasB/FasC | FasA | (+) FasX | fasBCA (TCS), ska (streptokinase), pilus operon, cpa (minor pilin protein). | Adhesion, motility and adherence | Unknown | [88,89,90] |
| CiaH | CiaR | (+) 5 csRNAs | comC (precursor of the competence stimulating peptide) | Competence, genomic plasticity | Unknown | [91] | |
| CovS | CovR | (+) RivX | possibly mga (master TR) | Mga regulates genes important for virulence in the host | Unknown | [92] | |
| Clostridium perfringens | VirS | VirR | (+) VR-RNA VirT, VirU | VR-RNA: plc (Phospholipase C), colA (κ-toxin) | Expression of toxins | [93,94,95,96,97] | |
| Pathogen | sRNA Regulating TCS | Regulated TCS | References |
|---|---|---|---|
| Gram-negative Bacteria | |||
| Escherichia coli | RybC | DpiA-DpiB | [193] |
| E. coli | SdsN | NarQ-NarP | [194] |
| E. coli, Salmonella | GcvB, MicA | PhoP-PhoQ | [195,196] |
| Gram-positive Bacteria | |||
| Staphylococcus aureus | psm-mec | AgrA-AgrC | [150] |
| Clostridioides difficile | Cd2-2 | CmrR-CmrS-CmrT | [182] |
| Streptococcus pneumoniae | srn206 | ComD-ComE | [170] |
| Enterococcus faecalis, Listeria monocytogenes | EutX, Rli55 | EutVW | [183,184] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piattelli, E.; Peltier, J.; Soutourina, O. Interplay between Regulatory RNAs and Signal Transduction Systems during Bacterial Infection. Genes 2020, 11, 1209. https://doi.org/10.3390/genes11101209
Piattelli E, Peltier J, Soutourina O. Interplay between Regulatory RNAs and Signal Transduction Systems during Bacterial Infection. Genes. 2020; 11(10):1209. https://doi.org/10.3390/genes11101209
Chicago/Turabian StylePiattelli, Emma, Johann Peltier, and Olga Soutourina. 2020. "Interplay between Regulatory RNAs and Signal Transduction Systems during Bacterial Infection" Genes 11, no. 10: 1209. https://doi.org/10.3390/genes11101209
APA StylePiattelli, E., Peltier, J., & Soutourina, O. (2020). Interplay between Regulatory RNAs and Signal Transduction Systems during Bacterial Infection. Genes, 11(10), 1209. https://doi.org/10.3390/genes11101209