Evidence for Allele-Specific Levels of Enhanced Susceptibility of Wheat mlo Mutants to the Hemibiotrophic Fungal Pathogen Magnaporthe oryzae pv. Triticum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Bgt Infection Assays
2.3. Z. tritici Infection Assays
2.4. MoT Infection Assays
2.5. Scoring of Spontaneous Callose Deposition
2.6. Assessment of Leaf Chlorosis/Necrosis
2.7. Statistical Analysis of The Data
3. Results
3.1. Tamlo Lines Show Allele-Dependent Levels of Powdery Mildew Resistance
3.2. Tamlo Triple-Mutants Show Unaltered Infection by The Fungal Pathogen Zymoseptoria tritici
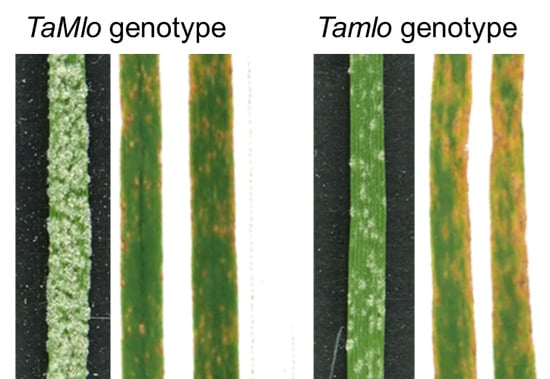
3.3. Tamlo Triple-Mutant Lines Show Allele-Dependent Levels of Enhanced Susceptibility to The Fungal Pathogen Magnaporthe oryzae pv. Triticum
3.4. Tamlo Triple-Mutants Do Not Exhibit Signs of Early Leaf Senescence
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | analysis of variance |
| BCI | backcross Ingrid |
| Bgt | Blumeria graminis f.sp. tritici |
| cv. | cultivar |
| dpi | days post inoculation |
| hpi | hours post inoculation |
| Mlo | Mildew locus o |
| MoT | Magnaporthe oryzae pv. Triticum |
| TALEN | Transcription activator-like effector nuclease |
| TILLING | Targeted induced local lesions in genomes |
References
- Shiferaw, B.; Smale, M.; Braun, H.-J.; Duveiller, E.; Reynolds, M.; Muricho, G. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Sec. 2013, 5, 291–317. [Google Scholar] [CrossRef] [Green Version]
- Frantzeskakis, L.; Di Pietro, A.; Rep, M.; Schirawski, J.; Wu, C.-H.; Panstruga, R. Rapid evolution in plant-microbe interactions—A molecular genomics perspective. New Phytol. 2020, 225, 1134–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glawe, D.A. The powdery mildews: A review of the world’s most familiar (yet poorly known) plant pathogens. Annu. Rev. Phytopathol. 2008, 46, 27–51. [Google Scholar] [CrossRef] [PubMed]
- Panstruga, R.; Schulze-Lefert, P. Live and let live: Insights into powdery mildew disease and resistance. Mol. Plant Pathol. 2002, 3, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Troch, V.; Audenaert, K.; Wyand, R.A.; Haesaert, G.; Höfte, M.; Brown, J.K. Formae speciales of cereal powdery mildew: Close or distant relatives? Mol. Plant Pathol. 2014, 15, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.A.; Xue, F.; Wang, C.; Ji, W. Powdery mildew resistance genes in wheat: Identification and genetic analysis. J. Mol. Biol. Res. 2011, 1, 20–39. [Google Scholar] [CrossRef] [Green Version]
- Jørgensen, J.H. Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 1994, 13, 97–119. [Google Scholar] [CrossRef]
- Jørgensen, J.H. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 1992, 63, 141–152. [Google Scholar] [CrossRef]
- Piffanelli, P.; Ramsay, L.; Waugh, R.; Benabdelmouna, A.; D’Hont, A.; Hollricher, K.; Jørgensen, J.H.; Schulze-Lefert, P.; Panstruga, R. A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature 2004, 430, 887–891. [Google Scholar] [CrossRef] [Green Version]
- Kusch, S.; Panstruga, R. mlo-based resistance: An apparently universal “weapon” to defeat powdery mildew disease. Mol. Plant-Microbe Interact. 2017, 30, 179–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consonni, C.; Humphry, M.E.; Hartmann, H.A.; Livaja, M.; Durner, J.; Westphal, L.; Vogel, J.; Lipka, V.; Kemmerling, B.; Schulze-Lefert, P.; et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 2006, 38, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Lyngkjær, M.F.; Newton, A.C.; Atzema, J.L.; Baker, S.J. The barley mlo gene: An important powdery mildew resistance source. Agronomie 2000, 20, 745–756. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.K.M. Durable resistance of crops to disease: A Darwinian perspective. Annu. Rev. Phytopathol. 2015, 53, 513–539. [Google Scholar] [CrossRef] [PubMed]
- Acevedo-Garcia, J.; Kusch, S.; Panstruga, R. Magical mystery tour—MLO proteins in plant immunity and beyond. New Phytol. 2014, 204, 273–281. [Google Scholar] [CrossRef]
- Elliott, C.; Zhou, F.S.; Spielmeyer, W.; Panstruga, R.; Schulze-Lefert, P. Functional conservation of wheat and rice Mlo orthologs in defense modulation to the powdery mildew fungus. Mol. Plant-Microbe Interact. 2002, 15, 1069–1077. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.-L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Spencer, D.; Thieron, H.; Reinstadler, A.; Hammond-Kosack, K.; Phillips, A.L.; Panstruga, R. mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 2017, 15, 367–378. [Google Scholar] [CrossRef]
- Ingvardsen, C.R.; Massange-Sánchez, J.A.; Borum, F.; Uauy, C.; Gregersen, P.L. Development of mlo-based resistance in tetraploid wheat against wheat powdery mildew. Theor. Appl. Genet. 2019, 132, 3009–3022. [Google Scholar] [CrossRef]
- Wolter, M.; Hollricher, K.; Salamini, F.; Schulze-Lefert, P. The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defense mimic phenotype. Mol. Gen. Genet. 1993, 239, 122–128. [Google Scholar] [CrossRef]
- Schwarzbach, E. The pleiotropic effects of the ml-o gene and their implications in breeding. In Barley Genetics III; Karl Thiemig: München, Germany, 1976; pp. 440–445. [Google Scholar]
- Piffanelli, P.; Zhou, F.S.; Casais, C.; Orme, J.; Jarosch, B.; Schaffrath, U.; Collins, N.C.; Panstruga, R.; Schulze-Lefert, P. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002, 129, 1076–1085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consonni, C.; Bednarek, P.; Humphry, M.; Francocci, F.; Ferrari, S.; Harzen, A.; van Themaat, E.V.L.; Panstruga, R. Tryptophan-derived metabolites are required for antifungal defense in the Arabidopsis mlo2 mutant. Plant Physiol. 2010, 152, 1544–1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.L.; Pavan, S.; Zheng, Z.; Zappel, N.F.; Reinstädler, A.; Lotti, C.; de Giovanni, C.; Ricciardi, L.; Lindhout, P.; Visser, R.; et al. Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of Mlo function. Mol. Plant-Microbe Interact. 2008, 21, 30–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphry, M.; Reinstädler, A.; Ivanov, S.; Bisseling, T.; Panstruga, R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol. Plant Pathol. 2011, 12, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Jarosch, B.; Kogel, K.H.; Schaffrath, U. The ambivalence of the barley Mlo locus: Mutations conferring resistance against powdery mildew (Blumeria graminis f. sp. hordei) enhance susceptibility to the rice blast fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 1999, 12, 508–514. [Google Scholar] [CrossRef] [Green Version]
- Inoue, Y.; Vy, T.T.P.; Yoshida, K.; Asano, H.; Mitsuoka, C.; Asuke, S.; Anh, V.L.; Cumagun, C.J.R.; Chuma, I.; Terauchi, R.; et al. Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science 2017, 357, 80–83. [Google Scholar] [CrossRef] [Green Version]
- Gladieux, P.; Condon, B.; Ravel, S.; Soanes, D.; Maciel, J.L.N.; Nhani, A.; Chen, L.; Terauchi, R.; Lebrun, M.-H.; Tharreau, D.; et al. Gene flow between divergent cereal- and grass-specific lineages of the rice blast fungus Magnaporthe oryzae. mBio 2018, 9, e01219-17. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.T.; Croll, D.; Gladieux, P.; Soanes, D.M.; Persoons, A.; Bhattacharjee, P.; Hossain, M.S.; Gupta, D.R.; Rahman, M.M.; Mahboob, M.G.; et al. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 2016, 14, 84. [Google Scholar] [CrossRef] [Green Version]
- Ceresini, P.C.; Castroagudín, V.L.; Rodrigues, F.Á.; Rios, J.A.; Aucique-Pérez, C.E.; Moreira, S.I.; Croll, D.; Alves, E.; de Carvalho, G.; Maciel, J.L.N.; et al. Wheat blast: From its origins in South America to its emergence as a global threat. Mol. Plant Pathol. 2019, 20, 155–172. [Google Scholar] [CrossRef]
- Valent, B.; Farman, M.; Tosa, Y.; Begerow, D.; Fournier, E.; Gladieux, P.; Islam, M.T.; Kamoun, S.; Kemler, M.; Kohn, L.M.; et al. Pyricularia graminis-tritici is not the correct species name for the wheat blast fungus: Response to Ceresini et al. (MPP 20:2). Mol. Plant Pathol. 2019, 20, 173–179. [Google Scholar] [CrossRef] [Green Version]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; van Daelen, R.; van der Lee, T.; Diergaarde, P.; Groenendijk, J.; et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef] [Green Version]
- Hinze, K.; Thompson, R.D.; Ritter, E.; Salamini, F.; Schulze-Lefert, P. Restriction fragment length polymorphism-mediated targeting of the ml-o resistance locus in barley (Hordeum vulgare). Proc. Natl. Acad. Sci. USA 1991, 88, 3691–3695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thygesen, K.; Jørgensen, L.N.; Jensen, K.S.; Munk, L. Spatial and temporal impact of fungicide spray strategies on fungicide sensitivity of Mycosphaerella graminicola in winter wheat. Eur. J. Plant Pathol. 2009, 123, 435–447. [Google Scholar] [CrossRef]
- Grandaubert, J.; Dutheil, J.Y.; Stukenbrock, E.H. The genomic determinants of adaptive evolution in a fungal pathogen. Evol. Lett. 2019, 3, 299–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habig, M.; Quade, J.; Stukenbrock, E.H. Forward genetics approach reveals host genotype-dependent importance of accessory chromosomes in the fungal wheat pathogen Zymoseptoria tritici. mBio 2017, 8, e01919-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, E.L.; Hagerty, C.H.; Mikaberidze, A.; Mundt, C.C.; Zhong, Z.; McDonald, B.A. An improved method for measuring quantitative resistance to the wheat pathogen Zymoseptoria tritici using high-throughput automated image analysis. Phytopathology 2016, 106, 782–788. [Google Scholar] [CrossRef] [Green Version]
- Faivre-Rampant, O.; Thomas, J.; Allègre, M.; Morel, J.-B.; Tharreau, D.; Nottéghem, J.-L.; Lebrun, M.-H.; Schaffrath, U.; Piffanelli, P. Characterization of the model system rice--Magnaporthe for the study of nonhost resistance in cereals. New Phytol. 2008, 180, 899–910. [Google Scholar] [CrossRef]
- Tanaka, M.; Nakayashiki, H.; Tosa, Y. Population structure of Eleusine isolates of Pyricularia oryzae and its evolutionary implications. J. Gen. Plant Pathol. 2009, 75, 173–180. [Google Scholar] [CrossRef]
- Strugala, R.; Delventhal, R.; Schaffrath, U. An organ-specific view on non-host resistance. Front. Plant Sci. 2015, 6, 526. [Google Scholar] [CrossRef] [Green Version]
- Delventhal, R.; Rajaraman, J.; Stefanato, F.L.; Rehman, S.; Aghnoum, R.; McGrann, G.R.D.; Bolger, M.; Usadel, B.; Hedley, P.E.; Boyd, L.; et al. A comparative analysis of nonhost resistance across the two Triticeae crop species wheat and barley. BMC Plant Biol. 2017, 17, 232. [Google Scholar] [CrossRef] [Green Version]
- Ulferts, S.; Delventhal, R.; Splivallo, R.; Karlovsky, P.; Schaffrath, U. Abscisic acid negatively interferes with basal defence of barley against Magnaporthe oryzae. BMC Plant Biol. 2015, 15, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, T.R.; Kang, I.H.; Wheeler, D.B.; Lindquist, R.A.; Papallo, A.; Sabatini, D.M.; Golland, P.; Carpenter, A.E. CellProfiler Analyst: Data exploration and analysis software for complex image-based screens. BMC Bioinform. 2008, 9, 482. [Google Scholar] [CrossRef] [Green Version]
- Haueisen, J.; Möller, M.; Eschenbrenner, C.J.; Grandaubert, J.; Seybold, H.; Adamiak, H.; Stukenbrock, E.H. Highly flexible infection programs in a specialized wheat pathogen. Ecol. Evol. 2019, 9, 275–294. [Google Scholar] [CrossRef] [PubMed]
- Zellerhoff, N.; Jarosch, B.; Groenewald, J.Z.; Crous, P.W.; Schaffrath, U. Nonhost resistance of barley is successfully manifested against Magnaporthe grisea and a closely related Pennisetum-infecting lineage but is overcome by Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2006, 19, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Jarosch, B.; Jansen, M.; Schaffrath, U. Acquired resistance functions in mlo barley, which is hypersusceptible to Magnaporthe grisea. Mol. Plant-Microbe Interact. 2003, 16, 107–114. [Google Scholar] [CrossRef] [Green Version]
- Jarosch, B.; Collins, N.C.; Zellerhoff, N.; Schaffrath, U. RAR1, ROR1, and the actin cytoskeleton contribute to basal resistance to Magnaporthe grisea in barley. Mol. Plant-Microbe Interact. 2005, 18, 397–404. [Google Scholar] [CrossRef] [Green Version]
- Habekuss, A.; Hentrich, W. Charakterisierung funktionell verschiedener ml-o-Mutanten durch Primärinfektion, Pustelwachstum, Inkubation und Befallsverlauf. Tag.-Ber. Akad. Landwirtsch.-Wiss. 1988, 272, 229–237. [Google Scholar]
- Hentrich, W. Multiple Allelie, Pleiotropie und züchterische Nutzung mehltauresistenter Mutanten des mlo-Locus der Gerste. Tag.-Ber. Akad. Landwirtsch.-Wiss. 1979, 175, 191–202. [Google Scholar]
- Hentrich, W.; Habekuss, A. Untersuchungen an heteroallelen Mutanten des ml-o Locus der Sommergerste. Schriftenreihe VAFB Thüringen 1993, 1/1993, 61–69. [Google Scholar]
- Sánchez-Vallet, A.; McDonald, M.C.; Solomon, P.S.; McDonald, B.A. Is Zymoseptoria tritici a hemibiotroph? Fungal Genet. Biol. 2015, 79, 29–32. [Google Scholar] [CrossRef]
- Fernandez, J.; Orth, K. Rise of a cereal killer: The biology of Magnaporthe oryzae biotrophic growth. Trends Microbiol. 2018, 26, 582–597. [Google Scholar] [CrossRef] [PubMed]
- Le Fevre, R.; O’Boyle, B.; Moscou, M.J.; Schornack, S. Colonization of barley by the broad-host hemibiotrophic pathogen Phytophthora palmivora uncovers a leaf development-dependent involvement of Mlo. Mol. Plant-Microbe Interact. 2016, 29, 385–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, J.; Hückelhoven, R.; Beckhove, U.; Nagarajan, S.; Kogel, K.H. A compromised Mlo pathway affects the response of barley to the necrotrophic fungus Bipolaris sorokiniana (teleomorph: Cochliobolus sativus) and its toxins. Phytopathology 2001, 91, 127–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansen, C.; von Wettstein, D.; Schäfer, W.; Kogel, K.-H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofer, K.; Linkmeyer, A.; Textor, K.; Hückelhoven, R.; Hess, M. MILDEW LOCUS O mutation does not affect resistance to grain infections with Fusarium spp. and Ramularia collo-cygni. Phytopathology 2015, 105, 1214–1219. [Google Scholar] [CrossRef] [Green Version]
- McGrann, G.R.D.; Stavrinides, A.; Russell, J.; Corbitt, M.M.; Booth, A.; Chartrain, L.; Thomas, W.T.B.; Brown, J.K.M. A trade off between mlo resistance to powdery mildew and increased susceptibility of barley to a newly important disease, Ramularia leaf spot. J. Exp. Bot. 2014, 65, 1025–1037. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Lozano, J.M.; Gianinazzi, S.; Gianinazzi-Pearson, V. Genes involved in resistance to powdery mildew in barley differentially modulate root colonization by the mycorrhizal fungus Glomus mosseae. Mycorrhiza 1999, 9, 237–240. [Google Scholar] [CrossRef]
- Hilbert, M.; Novero, M.; Rovenich, H.; Mari, S.; Grimm, C.; Bonfante, P.; Zuccaro, A. MLO differentially regulates barley root colonization by beneficial endophytic and mycorrhizal fungi. Front. Plant Sci. 2020, 10, 9319. [Google Scholar] [CrossRef] [Green Version]
- Jacott, C.N.; Charpentier, M.; Murray, J.D.; Ridout, C.J. Mildew Locus O facilitates colonization by arbuscular mycorrhizal fungi in angiosperms. New Phytol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Acevedo-Garcia, J.; Gruner, K.; Reinstädler, A.; Kemen, A.; Kemen, E.; Cao, L.; Takken, F.L.W.; Reitz, M.U.; Schäfer, P.; O’Connell, R.J.; et al. The powdery mildew-resistant Arabidopsis mlo2 mlo6 mlo12 triple mutant displays altered infection phenotypes with diverse types of phytopathogens. Sci. Rep. 2017, 7, 27. [Google Scholar] [CrossRef]
- Peterhänsel, C.; Freialdenhoven, A.; Kurth, J.; Kolsch, R.; Schulze-Lefert, P. Interaction analyses of genes required for resistance responses to powdery mildew in barley reveal distinct pathways leading to leaf cell death. Plant Cell 1997, 9, 1397–1409. [Google Scholar] [CrossRef] [PubMed]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gruner, K.; Esser, T.; Acevedo-Garcia, J.; Freh, M.; Habig, M.; Strugala, R.; Stukenbrock, E.; Schaffrath, U.; Panstruga, R. Evidence for Allele-Specific Levels of Enhanced Susceptibility of Wheat mlo Mutants to the Hemibiotrophic Fungal Pathogen Magnaporthe oryzae pv. Triticum. Genes 2020, 11, 517. https://doi.org/10.3390/genes11050517
Gruner K, Esser T, Acevedo-Garcia J, Freh M, Habig M, Strugala R, Stukenbrock E, Schaffrath U, Panstruga R. Evidence for Allele-Specific Levels of Enhanced Susceptibility of Wheat mlo Mutants to the Hemibiotrophic Fungal Pathogen Magnaporthe oryzae pv. Triticum. Genes. 2020; 11(5):517. https://doi.org/10.3390/genes11050517
Chicago/Turabian StyleGruner, Katrin, Tobias Esser, Johanna Acevedo-Garcia, Matthias Freh, Michael Habig, Roxana Strugala, Eva Stukenbrock, Ulrich Schaffrath, and Ralph Panstruga. 2020. "Evidence for Allele-Specific Levels of Enhanced Susceptibility of Wheat mlo Mutants to the Hemibiotrophic Fungal Pathogen Magnaporthe oryzae pv. Triticum" Genes 11, no. 5: 517. https://doi.org/10.3390/genes11050517
APA StyleGruner, K., Esser, T., Acevedo-Garcia, J., Freh, M., Habig, M., Strugala, R., Stukenbrock, E., Schaffrath, U., & Panstruga, R. (2020). Evidence for Allele-Specific Levels of Enhanced Susceptibility of Wheat mlo Mutants to the Hemibiotrophic Fungal Pathogen Magnaporthe oryzae pv. Triticum. Genes, 11(5), 517. https://doi.org/10.3390/genes11050517