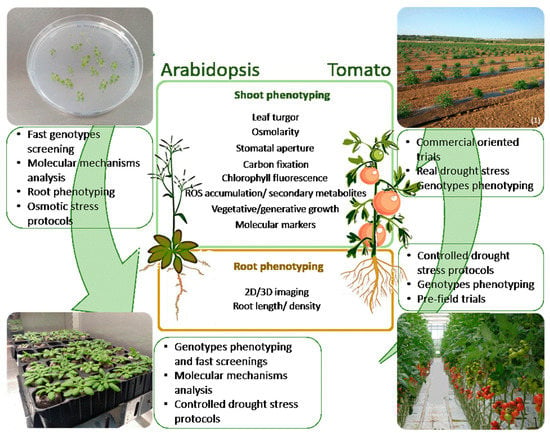
Phenotyping in Arabidopsis and Crops—Are We Addressing the Same Traits? A Case Study in Tomato
Abstract
:1. Introduction
2. The Multiple Facets of Drought
3. How Do Plants Cope with Drought? A Trait-Oriented Perspective
4. Drought Stress Protocols
5. Drought Stress Phenotyping
5.1. Leaf Turgor Drop
5.2. Osmolarity
5.3. Water Loss at the Leaf Level
5.4. Gas Exchange
5.5. Enhanced Chlorophyll Fluorescence
5.6. ROS and Leaf Secondary Metabolite Contents
5.7. Root Structure
5.8. Changes in Vegetative Growth
5.9. Changes in Generative Growth
5.10. Observing Stress through Marker Genes
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Iqbal, M.S.; Singh, A.K.; Ansari, M.I. Effect of Drought Stress on Crop Production. In New Frontiers in Stress Management for Durable Agriculture; Rakshit, A., Singh, H.B., Singh, A.K., Singh, U.S., Fraceto, L., Eds.; Springer: Singapore, 2020; pp. 35–47. ISBN 9789811513220. [Google Scholar]
- Gapp, K.S. The Universal Famine under Claudius. Harv. Theol. Rev. 1935, 28, 258–265. [Google Scholar] [CrossRef]
- Bourke, P.M.A. Emergence of Potato Blight, 1843–1846. Nature 1964, 203, 805–808. [Google Scholar] [CrossRef]
- Piguet, E.; Pécoud, A.; de Guchteneire, P. Migration and Climate Change: An Overview. Refug. Surv. Q. 2011, 30, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Maracchi, G.; Sirotenko, O.; Bindi, M. Impacts of Present and Future Climate Variability on Agriculture and Forestry in the Temperate Regions: Europe. Clim. Chang. 2005, 70, 117–135. [Google Scholar] [CrossRef]
- FAO. Water for Sustainable Food and Agriculture; FAO: Rome, Italy, 2016; ISBN 978-92-5-109977-3. [Google Scholar]
- Juenger, T.E. Natural variation and genetic constraints on drought tolerance. Curr. Opin. Plant Biol. 2013, 16, 274–281. [Google Scholar] [CrossRef]
- Kim, J.-I.; Kim, J.-Y. New era of precision plant breeding using genome editing. Plant Biotechnol. Rep. 2019, 13, 419–421. [Google Scholar] [CrossRef] [Green Version]
- Jankowicz-Cieslak, J.; Mba, C.; Till, B.J. Mutagenesis for Crop Breeding and Functional Genomics. In Biotechnologies for Plant Mutation Breeding: Protocols; Jankowicz-Cieslak, J., Tai, T.H., Kumlehn, J., Till, B.J., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 3–18. ISBN 978-3-319-45021-6. [Google Scholar]
- Page, D.R.; Grossniklaus, U. The art and design of genetic screens: Arabidopsis thaliana. Nat. Rev. Genet. 2002, 3, 124–136. [Google Scholar] [CrossRef]
- Weisburger, J.H. Lycopene and Tomato Products in Health Promotion. Exp. Biol. Med. 2002, 227, 924–927. [Google Scholar] [CrossRef] [PubMed]
- 100 Tomato Genome Sequencing Consortium; Aflitos, S.; Schijlen, E.; de Jong, H.; de Ridder, D.; Smit, S.; Finkers, R.; Wang, J.; Zhang, G.; Li, N.; et al. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. Cell Mol. Biol. 2014, 80, 136–148. [Google Scholar] [CrossRef] [Green Version]
- Ariizumi, T.; Shinozaki, Y.; Ezura, H. Genes that influence yield in tomato. Breed. Sci. 2013, 63, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.-K. Translational genomics and multi-omics integrated approaches as a useful strategy for crop breeding. Genes Genom. 2019, 41, 133–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Y.J.; Lee, T.; Lee, J.; Shim, S.; Jeong, H.; Satyawan, D.; Kim, M.Y.; Lee, S. Translational genomics for plant breeding with the genome sequence explosion. Plant Biotechnol. J. 2016, 14, 1057–1069. [Google Scholar] [CrossRef] [PubMed]
- Salentijn, E.M.J.; Pereira, A.; Angenent, G.C.; van der Linden, C.G.; Krens, F.; Smulders, M.J.M.; Vosman, B. Plant translational genomics: From model species to crops. Mol. Breed. 2007, 20, 1–13. [Google Scholar] [CrossRef]
- Nelissen, H.; Moloney, M.; Inzé, D. Translational research: From pot to plot. Plant Biotechnol. J. 2014, 12, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Novák, V. Physiological Drought—How to Quantify it? In Bioclimatology and 549 Natural Hazards; Strelcová, K., Matyas, C., Kleidon, A., Lapin, M., Matejka, F., Blazenec, M., Kvarenina, J., Holecy, J., Eds.; Springer: Dordrecht, The Netherland, 2009; pp. 89–95. [Google Scholar] [CrossRef]
- Rhodes, D.; Nadolska-Orczyk, A. Plant Stress Physiology. In eLS; American Cancer Society: Atlanta, GA, USA, 2001; ISBN 978-0-470-01590-2. [Google Scholar]
- Levitt, J. Responses of Plants to Environmental Stresses. Volume II. Water, Radiation, Salt, and Other Stresses; Academic Press: London, UK, 1971; ISBN 0-12-445502-6. [Google Scholar]
- Bouzid, M.; He, F.; Schmitz, G.; Häusler, R.E.; Weber, A.P.M.; Mettler-Altmann, T.; De Meaux, J. Arabidopsis species deploy distinct strategies to cope with drought stress. Ann. Bot. 2019, 124, 27–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, M.E.; Medina, V. Drought Adaptation Mechanisms Should Guide Experimental Design. Trends Plant Sci. 2016, 21, 639–647. [Google Scholar] [CrossRef] [Green Version]
- Wardlaw, I.F. Responses of Plants to Environmental Stresses; Levitt, J., Ed.; Academic Press: New York, NY, USA, 1972; Available online: https://science.sciencemag.org/content/177/4051/786.1 (accessed on 1 August 2020).
- Jensen, C.R.; Battilani, A.; Plauborg, F.; Psarras, G.; Chartzoulakis, K.; Janowiak, F.; Stikic, R.; Jovanovic, Z.; Li, G.; Qi, X.; et al. Deficit irrigation based on drought tolerance and root signalling in potatoes and tomatoes. Agric. Water Manag. 2010, 98, 403–413. [Google Scholar] [CrossRef]
- Landi, S.; De Lillo, A.; Nurcato, R.; Grillo, S.; Esposito, S. In-field study on traditional Italian tomato landraces: The constitutive activation of the ROS scavenging machinery reduces effects of drought stress. Plant Physiol. Biochem. 2017, 118, 150–160. [Google Scholar] [CrossRef]
- Vello, E.; Tomita, A.; Diallo, A.O.; Bureau, T.E. A Comprehensive Approach to Assess Arabidopsis Survival Phenotype in Water-Limited Condition Using a Non-invasive High-Throughput Phenomics Platform. Front. Plant Sci. 2015, 6, 1101. [Google Scholar] [CrossRef] [Green Version]
- Visentin, I.; Vitali, M.; Ferrero, M.; Zhang, Y.; Ruyter-Spira, C.; Novák, O.; Strnad, M.; Lovisolo, C.; Schubert, A.; Cardinale, F. Low levels of strigolactones in roots as a component of the systemic signal of drought stress in tomato. New Phytol. 2016, 212, 954–963. [Google Scholar] [CrossRef]
- Halperin, O.; Gebremedhin, A.; Wallach, R.; Moshelion, M. High-throughput physiological phenotyping and screening system for the characterization of plant-environment interactions. Plant J. 2017, 89, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Galdon-Armero, J.; Fullana-Pericas, M.; Mulet, P.A.; Conesa, M.A.; Martin, C.; Galmes, J. The ratio of trichomes to stomata is associated with water use efficiency in Solanum lycopersicum (tomato). Plant J. 2018, 96, 607–619. [Google Scholar] [CrossRef] [Green Version]
- Santaniello, A.; Scartazza, A.; Gresta, F.; Loreti, E.; Biasone, A.; Di Tommaso, D.; Piaggesi, A.; Perata, P. Ascophyllum nodosum Seaweed Extract Alleviates Drought Stress in Arabidopsis by Affecting Photosynthetic Performance and Related Gene Expression. Front. Plant Sci. 2017, 8, 1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz, D.; Litvin, A.G.; Fernandez, M.G.S. A cost-effective and customizable automated irrigation system for precise high-throughput phenotyping in drought stress studies. PLoS ONE 2018, 13, e0198546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frolov, A.; Bilova, T.; Paudel, G.; Berger, R.; Balcke, G.U.; Birkemeyer, C.; Wessjohann, L.A. Early responses of mature Arabidopsis thaliana plants to reduced water potential in the agar-based polyethylene glycol infusion drought model. J. Plant Physiol. 2017, 208, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Aazami, M.A.; Torabi, M.; Jalili, E. In Vitro response of promising tomato genotypes for tolerance to osmotic stress. Afr. J. Biotechnol. 2010, 9, 4014–4017. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Caballero, F.; Martínez, V.; Rubio, F. Disruption of the Arabidopsis thaliana Inward-Rectifier K+ Channel AKT1 Improves Plant Responses to Water Stress. Plant Cell Physiol. 2012, 53, 423–432. [Google Scholar] [CrossRef]
- Ali, N.; Schwarzenberg, A.; Yvin, J.-C.; Hosseini, S.A. Regulatory Role of Silicon in Mediating Differential Stress Tolerance Responses in Two Contrasting Tomato Genotypes under Osmotic Stress. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Amitai-Zeigerson, H.; Scolnik, P.A.; Bar-Zvi, D. Tomato Asr1 mRNA and protein are transiently expressed following salt stress, osmotic stress and treatment with abscisic acid. Plant Sci. 1995, 110, 205–213. [Google Scholar] [CrossRef]
- Jin, S.; Chen, C.C.S.; Plant, A.L. Regulation by ABA of osmotic-stress-induced changes in protein synthesis in tomato roots. Plant Cell Environ. 2000, 23, 51–60. [Google Scholar] [CrossRef]
- Visentin, I.; Pagliarani, C.; Deva, E.; Caracci, A.; Turečková, V.; Novák, O.; Lovisolo, C.; Schubert, A.; Cardinale, F. A novel strigolactone-miR156 module controls stomatal behaviour during drought recovery. Plant Cell Environ. 2020. [Google Scholar] [CrossRef]
- Virlouvet, L.; Ding, Y.; Fujii, H.; Avramova, Z.; Fromm, M. ABA signaling is necessary but not sufficient for RD29B transcriptional memory during successive dehydration stresses in Arabidopsis thaliana. Plant J. 2014, 79, 150–161. [Google Scholar] [CrossRef]
- Harb, A.; Krishnan, A.; Ambavaram, M.M.R.; Pereira, A. Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 2010, 154, 1254–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earl, H.J. A Precise Gravimetric Method for Simulating Drought Stress in Pot Experiments. Crop Sci. 2003, 43, 1868–1873. [Google Scholar] [CrossRef]
- Kishore, N.; Ashok kumar, K. Advanced Irrigation System using Arduino and Raspberry Pi as Centralized Server. Int. Res. J. Eng. Technol. 2019, 6, 4. [Google Scholar]
- Osmolovskaya, N.; Shumilina, J.; Kim, A.; Didio, A.; Grishina, T.; Bilova, T.; Keltsieva, O.A.; Zhukov, V.; Tikhonovich, I.; Tarakhovskaya, E.; et al. Methodology of Drought Stress Research: Experimental Setup and Physiological Characterization. Int. J. Mol. Sci. 2018, 19, 4089. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Nguyen, K.H.; Chu, H.D.; Ha, C.V.; Watanabe, Y.; Osakabe, Y.; Leyva-Gonzàlez, M.A.; Sato, M.; Toyooka, K.; Voges, L.; et al. The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana. PLoS Genet. 2017, 13, e1007076. [Google Scholar] [CrossRef] [Green Version]
- Marchin, R.M.; Ossola, A.; Leishman, M.R.; Ellsworth, D.S. A Simple Method for Simulating Drought Effects on Plants. Front. Plant Sci. 2020, 10, 1715. [Google Scholar] [CrossRef] [Green Version]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.-K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. Cell Mol. Biol. 2006, 45, 523–539. [Google Scholar] [CrossRef]
- de Ollas, C.; Segarra-Medina, C.; González-Guzmán, M.; Puertolas, J.; Gómez-Cadenas, A. A customizable method to characterize Arabidopsis thaliana transpiration under drought conditions. Plant Methods 2019, 15, 89. [Google Scholar] [CrossRef] [Green Version]
- van der Weele, C.M.; Spollen, W.G.; Sharp, R.E.; Baskin, T.I. Growth of Arabidopsis thaliana seedlings under water deficit studied by control of water potential in nutrient-agar media. J. Exp. Bot. 2000, 51, 1555–1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hohl, M.; Schopfer, P. Water Relations of Growing Maize Coleoptiles: Comparison between Mannitol and Polyethylene Glycol 6000 as External Osmotica for Adjusting Turgor Pressure. Plant Physiol. 1991, 95, 716–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slama, I.; Ghnaya, T.; Hessini, K.; Messedi, D.; Savouré, A.; Abdelly, C. Comparative study of the effects of mannitol and PEG osmotic stress on growth and solute accumulation in Sesuvium portulacastrum. Environ. Exp. Bot. 2007, 61, 10–17. [Google Scholar] [CrossRef]
- Koyama, R.; Itoh, H.; Kimura, S.; Morioka, A.; Uno, Y. Augmentation of Antioxidant Constituents by Drought Stress to Roots in Leafy Vegetables. HortTechnology 2012, 22, 121–125. [Google Scholar] [CrossRef] [Green Version]
- Ito, Y.; Katsura, K.; Maruyama, K.; Taji, T.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006, 47, 141–153. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Duursma, R.A.; Medlyn, B.E.; Kelly, J.W.G.; Prentice, I.C. How should we model plant responses to drought? An analysis of stomatal and non-stomatal responses to water stress. Agric. For. Meteorol. 2013, 182–183, 204–214. [Google Scholar] [CrossRef]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought “train” transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, 740. [Google Scholar] [CrossRef]
- Virlouvet, L.; Fromm, M. Physiological and transcriptional memory in guard cells during repetitive dehydration stress. New Phytol. 2015, 205, 596–607. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Yang, H.; Wang, L.; Liu, H.; Huo, H.; Zhang, C.; Liu, A.; Zhu, A.; Hu, J.; Lin, Y.; et al. Physiological and Transcriptome Analyses Reveal Short-Term Responses and Formation of Memory under Drought Stress in Rice. Front. Genet. 2019, 10, 55. [Google Scholar] [CrossRef] [Green Version]
- Costa, C.; Schurr, U.; Loreto, F.; Menesatti, P.; Carpentier, S. Plant Phenotyping Research Trends, a Science Mapping Approach. Front. Plant Sci. 2019, 9, 1933. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X.; et al. Unmanned Aerial Vehicle Remote Sensing for Field-Based Crop Phenotyping: Current Status and Perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef] [PubMed]
- Pieruschka, R.; Schurr, U. Plant Phenotyping: Past, Present, and Future. Available online: https://spj.sciencemag.org/plantphenomics/2019/7507131/?utm_campaign=trendmd-spj-test&utm_medium=cpc&utm_source=trendmd (accessed on 7 November 2019).
- Gosseau, F.; Blanchet, N.; Varès, D.; Burger, P.; Campergue, D.; Colombet, C.; Gody, L.; Liévin, J.-F.; Mangin, B.; Tison, G.; et al. Heliaphen, an Outdoor High-Throughput Phenotyping Platform for Genetic Studies and Crop Modeling. Front. Plant Sci. 2019, 9, 1908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.M.; Driever, S.M.; Heuvelink, E.; Rüger, S.; Zimmermann, U.; de Gelder, A.; Marcelis, L.F.M. Evaluation of diel patterns of relative changes in cell turgor of tomato plants using leaf patch clamp pressure probes. Physiol. Plant 2012, 146, 439–447. [Google Scholar] [CrossRef]
- Ache, P.; Bauer, H.; Kollist, H.; Al-Rasheid, K.A.S.; Lautner, S.; Hartung, W.; Hedrich, R. Stomatal action directly feeds back on leaf turgor: New insights into the regulation of the plant water status from non-invasive pressure probe measurements. Plant J. 2010, 62, 1072–1082. [Google Scholar] [CrossRef]
- Rose, J.C.; Paulus, S.; Kuhlmann, H. Accuracy Analysis of a Multi-View Stereo Approach for Phenotyping of Tomato Plants at the Organ Level. Sensors 2015, 15, 9651–9665. [Google Scholar] [CrossRef] [Green Version]
- Aghaie, P.; Hosseini Tafreshi, S.A.; Ebrahimi, M.A.; Haerinasab, M. Tolerance evaluation and clustering of fourteen tomato cultivars grown under mild and severe drought conditions. Sci. Hortic. 2018, 232, 1–12. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Zhang, Q.; Liu, Q.; Zhai, H.; Zhao, N.; He, S. An AP2/ERF gene, IbRAP2-12, from sweetpotato is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Sci. 2019, 281, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, K.; Zheng, Y.; Wang, Y.; Wang, J.; Liao, H. Disruption of AtWNK8 Enhances Tolerance of Arabidopsis to Salt and Osmotic Stresses via Modulating Proline Content and Activities of Catalase and Peroxidase. Int. J. Mol. Sci. 2013, 14, 7032–7047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Ortega, W.M.; Martínez, V.; Nieves, M.; Simón, I.; Lidón, V.; Fernandez-Zapata, J.C.; Martinez-Nicolas, J.J.; Cámara-Zapata, J.M.; García-Sánchez, F. Agricultural and Physiological Responses of Tomato Plants Grown in Different Soilless Culture Systems with Saline Water under Greenhouse Conditions. Sci. Rep. 2019, 9, 6733. [Google Scholar] [CrossRef] [PubMed]
- Verslues, P.E.; Bray, E.A. LWR1 and LWR2 Are Required for Osmoregulation and Osmotic Adjustment in Arabidopsis. Plant Physiol. 2004, 136, 2831–2842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merlot, S.; Mustilli, A.-C.; Genty, B.; North, H.; Lefebvre, V.; Sotta, B.; Vavasseur, A.; Giraudat, J. Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J. 2002, 30, 601–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leinonen, I.; Jones, H.G. Combining thermal and visible imagery for estimating canopy temperature and identifying plant stress. J. Exp. Bot. 2004, 55, 1423–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuromori, T.; Sugimoto, E.; Shinozaki, K. Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J. 2011, 67, 885–894. [Google Scholar] [CrossRef] [PubMed]
- Caird, M.A.; Richards, J.H.; Hsiao, T.C. Significant transpirational water loss occurs throughout the night in field-grown tomato. Funct. Plant Biol. 2007, 34, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Jung, S. Variation in antioxidant metabolism of young and mature leaves of Arabidopsis thaliana subjected to drought. Plant Sci. 2004, 166, 459–466. [Google Scholar] [CrossRef]
- Takayama, K.; Nishina, H.; Iyoki, S.; Arima, S.; Hatou, K.; Ueka, Y.; Miyoshi, Y. Early detection of drought stress in tomato plants with chlorophyll fluorescence imaging–practical application of the speaking plant approach in a greenhouse. IFAC Proc. Vol. 2011, 44, 1785–1790. [Google Scholar] [CrossRef]
- Yao, J.; Sun, D.; Cen, H.; Xu, H.; Weng, H.; Yuan, F.; He, Y. Phenotyping of Arabidopsis Drought Stress Response Using Kinetic Chlorophyll Fluorescence and Multicolor Fluorescence Imaging. Front. Plant Sci. 2018, 9, 603. [Google Scholar] [CrossRef]
- Mishra, K.B.; Iannacone, R.; Petrozza, A.; Mishra, A.; Armentano, N.; La Vecchia, G.; Trtílek, M.; Cellini, F.; Nedbal, L. Engineered drought tolerance in tomato plants is reflected in chlorophyll fluorescence emission. Plant Sci. 2012, 182, 79–86. [Google Scholar] [CrossRef]
- Fichman, Y.; Miller, G.; Mittler, R. Whole-Plant Live Imaging of Reactive Oxygen Species. Mol. Plant 2019, 12, 1203–1210. [Google Scholar] [CrossRef] [Green Version]
- Ijaz, R.; Ejaz, J.; Gao, S.; Liu, T.; Imtiaz, M.; Ye, Z.; Wang, T. Overexpression of annexin gene AnnSp2, enhances drought and salt tolerance through modulation of ABA synthesis and scavenging ROS in tomato. Sci. Rep. 2017, 7, 12087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Seo, P.J.; Lee, H.-J.; Park, C.-M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012, 70, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Susič, N.; Žibrat, U.; Širca, S.; Strajnar, P.; Razinger, J.; Knapič, M.; Vončina, A.; Urek, G.; Gerič Stare, B. Discrimination between abiotic and biotic drought stress in tomatoes using hyperspectral imaging. Sens. Actuators B Chem. 2018, 273, 842–852. [Google Scholar] [CrossRef] [Green Version]
- Mishra, P.; Feller, T.; Schmuck, M.; Nicol, A.; Nordon, A. Early Detection of Drought Stress in Arabidopsis thaliana Utilsing a Portable Hyperspectral Imaging Setup. In Proceedings of the 2019 10th Workshop on Hyperspectral Imaging and Signal Processing: Evolution in Remote Sensing (WHISPERS), Amsterdam, The Netherlands, 24–26 September 2019; pp. 1–5. [Google Scholar]
- Matsuda, O.; Tanaka, A.; Fujita, T.; Iba, K. Hyperspectral Imaging Techniques for Rapid Identification of Arabidopsis Mutants with Altered Leaf Pigment Status. Plant Cell Physiol. 2012, 53, 1154–1170. [Google Scholar] [CrossRef] [Green Version]
- Nakabayashi, R.; Mori, T.; Saito, K. Alternation of flavonoid accumulation under drought stress in Arabidopsis thaliana. Plant Signal. Behav. 2014, 9, e29518. [Google Scholar] [CrossRef] [Green Version]
- Hosoi, F.; Nakabayashi, K.; Omasa, K. 3-D Modeling of Tomato Canopies Using a High-Resolution Portable Scanning Lidar for Extracting Structural Information. Sensors 2011, 11, 2166–2174. [Google Scholar] [CrossRef]
- Paulus, S.; Behmann, J.; Mahlein, A.-K.; Plümer, L.; Kuhlmann, H. Low-Cost 3D Systems: Suitable Tools for Plant Phenotyping. Sensors 2014, 14, 3001–3018. [Google Scholar] [CrossRef] [Green Version]
- Jin, Z.; Sun, L.; Yang, G.; Pei, Y. Hydrogen Sulfide Regulates Energy Production to Delay Leaf Senescence Induced by Drought Stress in Arabidopsis. Front. Plant Sci. 2018, 9, 1722. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Ding, G.; Yokawa, K.; Baluška, F.; Li, Q.-F.; Liu, Y.; Shi, W.; Liang, J.; Zhang, J. An improved agar-plate method for studying root growth and response of Arabidopsis thaliana. Sci. Rep. 2013, 3, 1273. [Google Scholar] [CrossRef] [Green Version]
- Alaguero-Cordovilla, A.; Gran-Gómez, F.; Tormos-Moltó, S.; Pérez-Pérez, J. Morphological Characterization of Root System Architecture in Diverse Tomato Genotypes during Early Growth. Int. J. Mol. Sci. 2018, 19, 3888. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, L.; Lobet, G.; Tocquin, P.; Périlleux, C. “Rhizoponics”: A novel hydroponic rhizotron for root system analyses on mature Arabidopsis thaliana plants. Plant Methods 2015, 11, 3. [Google Scholar] [CrossRef] [Green Version]
- Mairhofer, S.; Zappala, S.; Tracy, S.R.; Sturrock, C.; Bennett, M.; Mooney, S.J.; Pridmore, T. RooTrak: Automated Recovery of Three-Dimensional Plant Root Architecture in Soil from X-ray Microcomputed Tomography Images Using Visual Tracking. Plant Physiol. 2012, 158, 561–569. [Google Scholar] [CrossRef] [Green Version]
- Jofuku, K.D.; Omidyar, P.K.; Gee, Z.; Okamuro, J.K. Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. USA 2005, 102, 3117–3122. [Google Scholar] [CrossRef] [Green Version]
- Sivakumar, R.; Srividhya, S.R. Impact of drought on flowering, yield and quality parameters in diverse genotypes of tomato (Solanum lycopersicum L.). Adv. Hortic. Sci. 2016, 30, 3–12. [Google Scholar] [CrossRef]
- Hao, G.-P.; Zhang, X.-H.; Wang, Y.-Q.; Wu, Z.-Y.; Huang, C.-L. Nucleotide Variation in the NCED3 Region of Arabidopsis thaliana and its Association Study with Abscisic Acid Content under Drought Stress. J. Integr. Plant Biol. 2009, 51, 175–183. [Google Scholar] [CrossRef]
- Yu, W.; Zhao, R.; Wang, L.; Zhang, S.; Li, R.; Sheng, J.; Shen, L. ABA signaling rather than ABA metabolism is involved in trehalose-induced drought tolerance in tomato plants. Planta 2019, 250, 643–655. [Google Scholar] [CrossRef]
- Muñoz-Espinoza, V.A.; López-Climent, M.F.; Casaretto, J.A.; Gómez-Cadenas, A. Water Stress Responses of Tomato Mutants Impaired in Hormone Biosynthesis Reveal Abscisic Acid, Jasmonic Acid and Salicylic Acid Interactions. Front. Plant Sci. 2015, 6, 997. [Google Scholar] [CrossRef] [Green Version]
- Sussmilch, F.C.; Brodribb, T.J.; McAdam, S.A.M. Up-regulation of NCED3 and ABA biosynthesis occur within minutes of a decrease in leaf turgor but AHK1 is not required. J. Exp. Bot. 2017, 68, 2913–2918. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.-F.; Liu, J.-K.; Yang, F.-M.; Zhang, G.-Y.; Wang, D.; Zhang, L.; Ou, Y.-B.; Yao, Y.-A. The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol. Plant 2020, 168, 98–117. [Google Scholar] [CrossRef]
- Ma, Y.; Cao, J.; Chen, Q.; He, J.; Liu, Z.; Wang, J.; Li, X.; Yang, Y. The Kinase CIPK11 Functions as a Negative Regulator in Drought Stress Response in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2422. [Google Scholar] [CrossRef] [Green Version]
- Iovieno, P.; Punzo, P.; Guida, G.; Mistretta, C.; Van Oosten, M.J.; Nurcato, R.; Bostan, H.; Colantuono, C.; Costa, A.; Bagnaresi, P.; et al. Transcriptomic Changes Drive Physiological Responses to Progressive Drought Stress and Rehydration in Tomato. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Liu, N.; Virlouvet, L.; Riethoven, J.-J.; Fromm, M.; Avramova, Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 229. [Google Scholar] [CrossRef] [Green Version]
- Hichri, I.; Muhovski, Y.; Clippe, A.; Žižková, E.; Dobrev, P.I.; Motyka, V.; Lutts, S. SlDREB2, a tomato dehydration-responsive element-binding 2 transcription factor, mediates salt stress tolerance in tomato and Arabidopsis. Plant Cell Environ. 2016, 39, 62–79. [Google Scholar] [CrossRef]
- Berger, B.; Parent, B.; Tester, M. High-throughput shoot imaging to study drought responses. J. Exp. Bot. 2010, 61, 3519–3528. [Google Scholar] [CrossRef] [Green Version]
- Dornbusch, T.; Lorrain, S.; Kuznetsov, D.; Fortier, A.; Liechti, R.; Xenarios, I.; Fankhauser, C. Measuring the diurnal pattern of leaf hyponasty and growth in Arabidopsis—A novel phenotyping approach using laser scanning. Funct. Plant Biol. 2012, 39, 860. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Rodríguez, E.; Rubio-Wilhelmi, M.; Cervilla, L.M.; Blasco, B.; Rios, J.J.; Rosales, M.A.; Romero, L.; Ruiz, J.M. Genotypic differences in some physiological parameters symptomatic for oxidative stress under moderate drought in tomato plants. Plant Sci. 2010, 178, 30–40. [Google Scholar] [CrossRef]
- Montesinos-Pereira, D.; Barrameda-Medina, Y.; Romero, L.; Ruiz, J.M.; Sánchez-Rodríguez, E. Genotype differences in the metabolism of proline and polyamines under moderate drought in tomato plants. Plant Biol. 2014, 16, 1050–1057. [Google Scholar] [CrossRef]
- Sperdouli, I.; Moustakas, M. Interaction of proline, sugars, and anthocyanins during photosynthetic acclimation of Arabidopsis thaliana to drought stress. J. Plant Physiol. 2012, 169, 577–585. [Google Scholar] [CrossRef]
- Sagor, G.H.M.; Zhang, S.; Kojima, S.; Simm, S.; Berberich, T.; Kusano, T. Reducing Cytoplasmic Polyamine Oxidase Activity in Arabidopsis Increases Salt and Drought Tolerance by Reducing Reactive Oxygen Species Production and Increasing Defense Gene Expression. Front. Plant Sci. 2016, 7, 214. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, K.; Takahashi, Y.; Berberich, T.; Imai, A.; Takahashi, T.; Michael, A.J.; Kusano, T. A protective role for the polyamine spermine against drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 2007, 352, 486–490. [Google Scholar] [CrossRef]
- Hazarika, P.; Rajam, M.V. Biotic and abiotic stress tolerance in transgenic tomatoes by constitutive expression of S-adenosylmethionine decarboxylase gene. Physiol. Mol. Biol. Plants 2011, 17, 115–128. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.-Q.; Zhang, Q.-F.; Liu, J.-H.; Li, G.-H. Overexpression of PtADC confers enhanced dehydration and drought tolerance in transgenic tobacco and tomato: Effect on ROS elimination. Biochem. Biophys. Res. Commun. 2011, 413, 10–16. [Google Scholar] [CrossRef]
- Gitz, D.C.; Baker, J.T. Methods for Creating Stomatal Impressions Directly onto Archivable Slides. Agron. J. 2009, 101, 232–236. [Google Scholar] [CrossRef]
- Bhugra, S.; Mishra, D.; Anupama, A.; Chaudhury, S.; Lall, B.; Chugh, A.; Chinnusamy, V. Deep Convolutional Neural Networks Based Framework for Estimation of Stomata Density and Structure from Microscopic Images. In Computer Vision—ECCV 2018 Workshops; Leal-Taixé, L., Roth, S., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, The Netherland, 2019; Volume 11134, pp. 412–423. ISBN 978-3-030-11023-9. [Google Scholar]
- Jäger, K.; Fábián, A.; Tompa, G.; Deák, C.; Höhn, M.; Olmedilla, A.; Barnabás, B.; Papp, I. New phenotypes of the drought-tolerant cbp20 Arabidopsis thaliana mutant have changed epidermal morphology. Plant Biol. 2011, 13, 78–84. [Google Scholar] [CrossRef]
- Vadez, V.; Kholová, J.; Hummel, G.; Zhokhavets, U.; Gupta, S.K.; Hash, C.T. LeasyScan: A novel concept combining 3D imaging and lysimetry for high-throughput phenotyping of traits controlling plant water budget. J. Exp. Bot. 2015, 66, 5581–5593. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Kinoshita, T. Stomatal function has an element of hysteresis. New Phytol. 2015, 205, 455–457. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R. When Defense Pathways Collide. The Response of Arabidopsis to a Combination of Drought and Heat Stress. Plant Physiol. 2004, 134, 1683–1696. [Google Scholar] [CrossRef] [Green Version]
- Alcázar, R.; Planas, J.; Saxena, T.; Zarza, X.; Bortolotti, C.; Cuevas, J.; Bitrián, M.; Tiburcio, A.F.; Altabella, T. Putrescine accumulation confers drought tolerance in transgenic Arabidopsis plants over-expressing the homologous Arginine decarboxylase 2 gene. Plant Physiol. Biochem. 2010, 48, 547–552. [Google Scholar] [CrossRef]
- Baker, N.R. Chlorophyll Fluorescence: A Probe of Photosynthesis in Vivo. Annu. Rev. Plant Biol. 2008, 59, 89–113. [Google Scholar] [CrossRef] [Green Version]
- Tripodi, P.; Massa, D.; Venezia, A.; Cardi, T. Sensing Technologies for Precision Phenotyping in Vegetable Crops: Current Status and Future Challenges. Agronomy 2018, 8, 57. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Wu, Z.; Wang, X.; Rosenqvist, E.; Wang, Y.; Zhao, T.; Ottosen, C.-O. Evaluation of temperature stress tolerance in cultivated and wild tomatoes using photosynthesis and chlorophyll fluorescence. Hortic. Environ. Biotechnol. 2018, 59, 499–509. [Google Scholar] [CrossRef]
- Liu, N.; Jin, Z.; Wang, S.; Gong, B.; Wen, D.; Wang, X.; Wei, M.; Shi, Q. Sodic alkaline stress mitigation with exogenous melatonin involves reactive oxygen metabolism and ion homeostasis in tomato. Sci. Hortic. 2015, 181, 18–25. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Chivkunova, O.B.; Solovchenko, A.E.; Naqvi, K.R. Light absorption by anthocyanins in juvenile, stressed, and senescing leaves. J. Exp. Bot. 2008, 59, 3903–3911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Junker-Frohn, L.V.; Lück, M.; Schmittgen, S.; Wensing, J.; Carraresi, L.; Thiele, B.; Groher, T.; Reimer, J.J.; Bröring, S.; Noga, G.; et al. Tomato’s Green Gold: Bioeconomy Potential of Residual Tomato Leaf Biomass as a Novel Source for the Secondary Metabolite Rutin. ACS Omega 2019, 4, 19071–19080. [Google Scholar] [CrossRef] [Green Version]
- Arab, M.; Bahramian, B.; Schindeler, A.; Valtchev, P.; Dehghani, F.; McConchie, R. Extraction of phytochemicals from tomato leaf waste using subcritical carbon dioxide. Innov. Food Sci. Emerg. Technol. 2019, 57, 102204. [Google Scholar] [CrossRef]
- Ellenberger, J.; Siefen, N.; Krefting, P.; Schulze Lutum, J.-B.; Pfarr, D.; Remmel, M.; Schröder, L.; Röhlen-Schmittgen, S. Effect of UV Radiation and Salt Stress on the Accumulation of Economically Relevant Secondary Metabolites in Bell Pepper Plants. Agronomy 2020, 10, 142. [Google Scholar] [CrossRef] [Green Version]
- Koevoets, I.T.; Venema, J.H.; Elzenga, J.T.M.; Testerink, C. Roots Withstanding their Environment: Exploiting Root System Architecture Responses to Abiotic Stress to Improve Crop Tolerance. Front. Plant Sci. 2016, 7, 1335. [Google Scholar] [CrossRef] [Green Version]
- Pflugfelder, D.; Metzner, R.; van Dusschoten, D.; Reichel, R.; Jahnke, S.; Koller, R. Non-invasive imaging of plant roots in different soils using magnetic resonance imaging (MRI). Plant Methods 2017, 13, 102. [Google Scholar] [CrossRef]
- Nagel, K.A.; Putz, A.; Gilmer, F.; Heinz, K.; Fischbach, A.; Pfeifer, J.; Faget, M.; Blossfeld, S.; Ernst, M.; Dimaki, C.; et al. GROWSCREEN-Rhizo is a novel phenotyping robot enabling simultaneous measurements of root and shoot growth for plants grown in soil-filled rhizotrons. Funct. Plant Biol. 2012, 39, 891. [Google Scholar] [CrossRef] [Green Version]
- Wasson, A.P.; Nagel, K.A.; Tracy, S.; Watt, M. Beyond Digging: Noninvasive Root and Rhizosphere Phenotyping. Trends Plant Sci. 2020, 25, 119–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mairhofer, S.; Pridmore, T.; Johnson, J.; Wells, D.M.; Bennett, M.J.; Mooney, S.J.; Sturrock, C.J. X-ray Computed Tomography of Crop Plant Root Systems Grown in Soil: X-ray Computed Tomography of Root Systems. Curr. Protoc. Plant Biol. 2017, 2, 270–286. [Google Scholar] [CrossRef]
- Bouteillé, M.; Rolland, G.; Balsera, C.; Loudet, O.; Muller, B. Disentangling the Intertwined Genetic Bases of Root and Shoot Growth in Arabidopsis. PLoS ONE 2012, 7, e32319. [Google Scholar] [CrossRef] [Green Version]
- Watt, M.; Moosavi, S.; Cunningham, S.C.; Kirkegaard, J.A.; Rebetzke, G.J.; Richards, R.A. A rapid, controlled-environment seedling root screen for wheat correlates well with rooting depths at vegetative, but not reproductive, stages at two field sites. Ann. Bot. 2013, 112, 447–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riboni, M.; Robustelli Test, A.; Galbiati, M.; Tonelli, C.; Conti, L. ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6309–6322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soyk, S.; Müller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jiménez-Gómez, J.M.; Lippman, Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017, 49, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Onouchi, H.; Coupland, G. The regulation of flowering time of Arabidopsis in response to daylength. J. Plant Res. 1998, 111, 271–275. [Google Scholar] [CrossRef]
- Grandillo, S.; Ku, H.M.; Tanksley, S.D. Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theor. Appl. Genet. 1999, 99, 978–987. [Google Scholar] [CrossRef]
- Gallego-Giraldo, C.; Hu, J.; Urbez, C.; Gomez, M.D.; Sun, T.; Perez-Amador, M.A. Role of the gibberellin receptors GID1 during fruit-set in Arabidopsis. Plant J. 2014, 79, 1020–1032. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Kuromori, T.; Sato, H.; Shinozaki, K. Regulatory Gene Networks in Drought Stress Responses and Resistance in Plants. Adv. Exp. Med. Biol. 2018, 1081, 189–214. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Dong, W.; Zhang, J.; Guo, X.; Chen, J.; Wang, Z.; Lin, Z.; Tang, H.; Zhang, L. The Sequenced Angiosperm Genomes and Genome Databases. Front. Plant Sci. 2018, 9, 418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaerle, L.; Lenk, S.; Leinonen, I.; Jones, H.G.; Van Der Straeten, D.; Buschmann, C. Multi-sensor plant imaging: Towards the development of a stress-catalogue. Biotechnol. J. 2009, 4, 1152–1167. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics—Technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Tardieu, F.; Cabrera-Bosquet, L.; Pridmore, T.; Bennett, M. Plant Phenomics, From Sensors to Knowledge. Curr. Biol. 2017, 27, R770–R783. [Google Scholar] [CrossRef]
- Esposito, S.; Carputo, D.; Cardi, T.; Tripodi, P. Applications and Trends of Machine Learning in Genomics and Phenomics for Next-Generation Breeding. Plants 2019, 9, 34. [Google Scholar] [CrossRef] [Green Version]
- Tardieu, F. Virtual plants: Modelling as a tool for the genomics of tolerance to water deficit. Trends Plant Sci. 2003, 8, 9–14. [Google Scholar] [CrossRef]
- De Swaef, T.; Steppe, K. Linking stem diameter variations to sap flow, turgor and water potential in tomato. Funct. Plant Biol. 2010, 37, 429. [Google Scholar] [CrossRef]
- Yuan, H.; Cheung, C.Y.M.; Poolman, M.G.; Hilbers, P.A.J.; Riel, N.A.W. van A genome-scale metabolic network reconstruction of tomato (Solanum lycopersicum L.) and its application to photorespiratory metabolism. Plant J. 2016, 85, 289–304. [Google Scholar] [CrossRef] [Green Version]
- Zardilis, A.; Hume, A.; Millar, A.J. A multi-model framework for the Arabidopsis life cycle. J. Exp. Bot. 2019, 70, 2463–2477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearl, J.; Bareinboim, E. External Validity: From Do-Calculus to Transportability across Populations. Stat. Sci. 2014, 29, 579–595. [Google Scholar] [CrossRef]
| Stress Application Method | Growth Substrate | Advantages (+)/Disadvantages (−) | Phenotyping Suitability | Arabidopsis | Tomato |
|---|---|---|---|---|---|
| Water withholding | Soil (open or protected field) | (+) realistic drought conditions | All traits can be phenotyped, but root phenotyping can be unfeasible | NA | Landi et al., 2017 [25] |
| (+) best method for market-oriented phenotyping | |||||
| (−) other stresses such as salinity and heat can co-occur | |||||
| (−) not used/useful for Arabidopsis | |||||
| (−) strongly affected by weather conditions | |||||
| Soil (pot) | (+) quite close to commercial conditions | All phenotyping methods here described can be used, but root phenotyping needs appropriate apparatus (e.g., rhizotrons, x-ray tomography) | Vello et al., 2015 [26] | Visentin et al., 2016 [27] Halperin et al., 2017 [28] Galdon-Armero et al., 2018 [29] | |
| (+) suitable for every growth stage | |||||
| (−) influenced by environmental conditions | |||||
| (−) can be laborious | |||||
| (−) stress can be slow to occur | |||||
| Soil (pellet) | (+) as for pot protocols, but the limited size of pellets speeds up drought stress occurrence | All phenotyping methods described here can be used | Vello et al., 2015 [26] | NA | |
| (−) not used for tomato | |||||
| Inert substrate e.g., sand, vermiculite (pot) | (+) stress is reached faster than in soil-based protocols | All phenotyping techniques described here can be carried out | Santaniello et al., 2017 [30] | Takayama et al., 2011 [31] | |
| (+) easier to uproot plants | |||||
| (−) nutrient stress occurs together with water withholding, as plants are fertigated | |||||
| (−) more artificial than soil-based protocols | |||||
| Transfer to stressing substrate | Agar with low osmotic potential | (+) very reproducible | Phenotyping, especially for tomato, is limited to the first stages of plant growth (seedling stage). Very convenient for early screenings | Frolov et al., 2017 [32] | Aazami et al., 2010 [33] |
| (+) a wide range of stress intensities can be achieved | |||||
| (+) fast | |||||
| (+) sterile | |||||
| (−) far from naturally occurring conditions | |||||
| (−) depending on osmolyte nature, off-target effects can be a concern | |||||
| (−) suitable only for small/young plants | |||||
| (−) stomata dynamics hard to assess in very young plants | |||||
| Hydroponics-Osmotic stress | (+) very reproducible | All phenotyping techniques described here can be carried out. Very suitable for the description of precise kinetics. Absence of soil makes root phenotyping not always feasible | Nieves-Cordones et al., 2012 [34] | Ali et al., 2019 [35] Amitai-Ziegerson et al., 1995 [36] | |
| (+) fast | |||||
| (+) a wide range of stress intensities can be achieved by gradually increasing osmolyte concentration | |||||
| (−) artificial | |||||
| (−) depending on solute nature, off-target effects can be a concern | |||||
| (−) root growth is altered | |||||
| (−) need for a hydroponic apparatus | |||||
| Inert substrates-Osmotic stress | (+) reproducible | All phenotyping techniques described here can be carried out. Very good if precise kinetics are analyzed. | NA | Jin et al., 2000 [37] | |
| (+) fast | |||||
| (+) a wide range of stress intensities can be achieved by gradually increasing osmolyte concentration | |||||
| (+) cost-effective | |||||
| (−) artificial | |||||
| (−) depending on solute nature, off-target effects can be a concern | |||||
| Transfer to dry substrate | Inert substrate | (+) very fast | Due to very fast stress, only early responses can be studied. Root phenotyping is not convenient | NA | Visentin et al., 2020 [38] |
| (+) reproducible | |||||
| (−) very artificial | |||||
| (−) severe stress only | |||||
| (−) only early responses can be analyzed | |||||
| Uproot and let dehydrate | Inert substrate to no substrate | (+) very fast | Due to very fast stress, only early responses can be studied. Root phenotyping is not convenient | Virlouvet et al., 2014 [39] | NA |
| (+) reproducible | |||||
| (−) very artificial | |||||
| (−) severe stress only | |||||
| (−) only early responses can be analyzed |
| Physiological Reaction Monitored | Accessible Traits | Arabidopsis | Tomato |
|---|---|---|---|
| Leaf turgor drop | - Direct assessment (high-precision pressure probe) | Direct assessment: | Direct assessment: Lee et al., 2012 [62] |
| - Wilting (RGB-imaging) | Ache et al., 2010 [63] | ||
| - Drop in projected leaf area | Plant architecture (Light Detection and Ranging—LiDAR): | ||
| - Lower specific leaf area | Wilting (RGB-imaging): Bouzid et al., 2019 [21] | Rose et al., 2015 [64] | |
| - Relative water content | Projected leaf area: | ||
| de Ollas et al., 2019 [47] | |||
| Osmolarity increase | - proline quantification | Proline: | Proline: Aghaie et al., 2018 [65] |
| - osmolarity quantification | Li et al., 2019 [66] | Osmolarity: | |
| Zhang et al., 2013 [67] | Rodríguez-Ortega et al., 2019 [68] | ||
| Osmolarity: | |||
| Frolov et al., 2017 [32] | |||
| Versluis & Bray, 2004 [69] | |||
| Stomata closure | - Leaf temperature (by infrared thermography) | Infrared thermography: | Infrared |
| - Direct stomata aperture measurements (by microscopy; destructive) | Li et al., 2017 [44] | thermography: | |
| - Stomatal conductance (by porometer) | Merlot et al., 2002 [70] | Leinonen & Jones, 2004 [71] | |
| Kuromori et al., 2011 [72] | Porometer: | ||
| Microscopy: | Visentin et al., 2020 [38] | ||
| Virlouvet & Fromm, 2014 [55] | Caird et al., 2007 [73] | ||
| Microscopy: | |||
| Galdon-Armero et al., 2018 [29] | |||
| Lower carbon fixation | - Leaf gas exchange | Harb et al., 2010 [40] | Galdon-Armero et al., 2018 [29] |
| Enhanced chlorophyll fluorescence | - Hand-held devices to assess chlorophyll fluorescence | Hand-held device: | Imaging system (within crop stand): |
| - Fluorescence imaging (e.g., PAM imaging) | Jung, 2004 [74] | Takayama et al., 2011 [75] | |
| PAM imaging: | Imaging system: | ||
| Yao et al., 2018 [76] | (FluorCamFC1000-H) | ||
| Mishra et al., 2012 [77] | |||
| Higher concentrations of Reactive Oxygen Species (ROS) in the leaf | - Chemical staining and imaging: destructive or non destructive | Non-destructive chemical imaging: | Destructive chemical imaging: |
| Fichman et al., 2019 [78] | Ijaz et al., 2017 [79] | ||
| Destructive chemical imaging: | |||
| Lee et al., 2012 [80] | |||
| Higher concentrations of ROS-scavenging secondary metabolites (e.g., flavonoids, anthocyanins, carotenoids) | - Hand-held devices for accessing specific leaf compounds (e.g., Dualex, Multiplex, FieldSpec) | Hyperspectral imaging: | Hyperspectral imaging: Susič et al., 2018 [81] |
| - Hyperspectral imaging | Mishra et al., 2019 [82] | Metabolomics: Ali et al., 2018 [35] | |
| - Full metabolic profiling (destructive) | Matsuda et al., 2012 [83] | ||
| Metabolomics: | |||
| Nakabayashi et al., 2014 [84] | |||
| Changes in vegetative growth | - RGB-Imaging: lower projected leaf area, compact habitus | RGB-Imaging: | LiDAR: Hosoi et al., 2011 [85] |
| - Lower fresh and dry mass | Ollas et al., 2019 [47] | 3D point clouds: Paulus et al., 2014 [86] | |
| - Lower specific leaf area | Senescence: Jin et al., 2018 [87] | Trichomes: Galdon-Armero et al., 2018 [29] | |
| - Slowed longitudinal growth of individual leaves | |||
| - Senescence | |||
| Changes in root growth | - 2D features | Xu et al., 2013 [88] | Alaguero-Cordovilla et al., 2018 [89] |
| - 3D features | Mathieu et al., 2015 [90] | Mairhofer et al., 2012 [91] | |
| Changes in generative growth | - Earlier fruit set | Seed mass and yield: Jofuku et al., 2005 [92] | Flowering and yield: Sivakumar et al., 2016 [93] |
| - Lower fruit weight | |||
| - Higher number of non-marketable fruits | |||
| - Lower overall yield | |||
| Molecular markers | - 9-Cis-Epoxycarotenoid Deoxygenase | AtNCED3: | SlNCED1, SlNCED2: |
| NCED | Hao et al., 2009 [94] | Yu et al., 2019 [95] | |
| Sussmilch, 2017 | Muoz-Espinoza et al., 2015 [96] | ||
| [97] | |||
| SlRD29: | |||
| - Responsive to dehydration 29 | AtRD29B: | Gao et al., 2020 [98] | |
| RD29 | Ma et al., 2019 [99] | Iovieno et al., 2016 [100] | |
| Virlouvet et al., 2014 [39] | |||
| NA | |||
| - Homeobox protein 6 | HB6: | ||
| HB6 | Ding et al., 2013 [101] | ||
| Harb et al., 2010 [40] | |||
| - Solyc02g084850 | (Unpublished data) | ||
| NA | |||
| - Dehydration-responsive Element- Binding protein 2 | SlDREB2: | ||
| DREB2 | AtDREB2A: | Gao et al., 2020 [98] | |
| Ma et al., 2019 [99] | Hichri et al., 2016 [102] | ||
| Harb et al., 2010 [40] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korwin Krukowski, P.; Ellenberger, J.; Röhlen-Schmittgen, S.; Schubert, A.; Cardinale, F. Phenotyping in Arabidopsis and Crops—Are We Addressing the Same Traits? A Case Study in Tomato. Genes 2020, 11, 1011. https://doi.org/10.3390/genes11091011
Korwin Krukowski P, Ellenberger J, Röhlen-Schmittgen S, Schubert A, Cardinale F. Phenotyping in Arabidopsis and Crops—Are We Addressing the Same Traits? A Case Study in Tomato. Genes. 2020; 11(9):1011. https://doi.org/10.3390/genes11091011
Chicago/Turabian StyleKorwin Krukowski, Paolo, Jan Ellenberger, Simone Röhlen-Schmittgen, Andrea Schubert, and Francesca Cardinale. 2020. "Phenotyping in Arabidopsis and Crops—Are We Addressing the Same Traits? A Case Study in Tomato" Genes 11, no. 9: 1011. https://doi.org/10.3390/genes11091011
APA StyleKorwin Krukowski, P., Ellenberger, J., Röhlen-Schmittgen, S., Schubert, A., & Cardinale, F. (2020). Phenotyping in Arabidopsis and Crops—Are We Addressing the Same Traits? A Case Study in Tomato. Genes, 11(9), 1011. https://doi.org/10.3390/genes11091011