Transcriptomic Analysis of a Diabetic Skin-Humanized Mouse Model Dissects Molecular Pathways Underlying the Delayed Wound Healing Response
Abstract
:1. Introduction
2. Materials and Methods
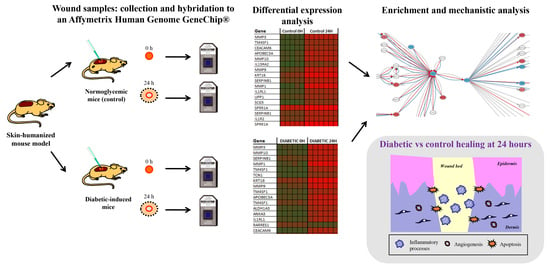
2.1. Wound Healing Experimental Design in the Diabetic Skin Humanized Mouse Model
2.2. RNA Extraction
2.3. Microarray Analysis
2.4. Data Processing and Statistical Analysis
2.5. Pathway Activity Analysis
3. Results
3.1. Exploratory Microarray Data Analysis
3.2. Differential Expression and Functional Enrichment Analysis of the Diabetes-Induced Skin-Humanized Mouse Model
3.3. Differential Expression and Functional Enrichment Analysis of the Wound Healing Process in the Diabetes-Induced Skin-Humanized Mouse Model
3.3.1. Common Transcriptomic Response to Wound Healing in Both Control and Diabetic Mice
3.3.2. Different Transcriptomic Response to Wound Healing in Control and Diabetic Mice
3.4. Mechanistic Signaling Pathway Analysis of the Wound Healing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, D.G.; Boulton, A.J.M.; Bus, S.A. Diabetic Foot Ulcers and Their Recurrence. N. Engl. J. Med. 2017, 376, 2367–2375. [Google Scholar] [CrossRef] [PubMed]
- Falanga, V. Wound healing and its impairment in the diabetic foot. Lancet 2005, 366, 1736–1743. [Google Scholar] [CrossRef]
- Blakytny, R.; Jude, E. The molecular biology of chronic wounds and delayed healing in diabetes. Diabet Med. 2006, 23, 594–608. [Google Scholar] [CrossRef] [PubMed]
- Brem, H.; Tomic-Canic, M. Cellular and molecular basis of wound healing in diabetes. J. Clin. Investig. 2007, 117, 1219–1222. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.; Srivastava, S.; Singh, M.R.; Singh, D. Mechanistic insight into diabetic wounds: Pathogenesis, molecular targets and treatment strategies to pace wound healing. Biomed. Pharm. 2019, 112, 108615. [Google Scholar] [CrossRef] [PubMed]
- O’Loughlin, A.; McIntosh, C.; Dinneen, S.F.; O’Brien, T. Review paper: Basic concepts to novel therapies: A review of the diabetic foot. Int. J. Low. Extrem. Wounds 2010, 9, 90–102. [Google Scholar] [CrossRef]
- Rees, D.A.; Alcolado, J.C. Animal models of diabetes mellitus. Diabet. Med. 2005, 22, 359–370. [Google Scholar] [CrossRef]
- Ansell, D.M.; Holden, K.A.; Hardman, M.J. Animal models of wound repair: Are they cutting it? Exp. Dermatol. 2012, 21, 581–585. [Google Scholar] [CrossRef]
- Grada, A.; Mervis, J.; Falanga, V. Research Techniques Made Simple: Animal Models of Wound Healing. J. Investig. Dermatol. 2018, 138, 2095–2105.e2091. [Google Scholar] [CrossRef] [Green Version]
- Del Rio, M.; Larcher, F.; Serrano, F.; Meana, A.; Muñoz, M.; Garcia, M.; Muñoz, E.; Martin, C.; Bernad, A.; Jorcano, J.L. A preclinical model for the analysis of genetically modified human skin in vivo. Hum. Gene Ther. 2002, 13, 959–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llames, S.G.; Del Rio, M.; Larcher, F.; García, E.; García, M.; Escamez, M.J.; Jorcano, J.L.; Holguín, P.; Meana, A. Human plasma as a dermal scaffold for the generation of a completely autologous bioengineered skin. Transplantation 2004, 77, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Escamez, M.; Martínez-Santamaría, L.; Garcia, M.; Guerrero-Aspizua, S.; Carretero, M.; Larcher, F.; Meana, A.; del Río, M. Bioengineered Skin. In Skin Biopsy: Perspectives; Khopkar, U., Ed.; InTech: London, UK, 2011; pp. 261–296. [Google Scholar] [CrossRef]
- Luchetti, M.M.; Moroncini, G.; Jose Escamez, M.; Svegliati Baroni, S.; Spadoni, T.; Grieco, A.; Paolini, C.; Funaro, A.; Avvedimento, E.V.; Larcher, F.; et al. Induction of Scleroderma Fibrosis in Skin-Humanized Mice by Administration of Anti-Platelet-Derived Growth Factor Receptor Agonistic Autoantibodies. Arthritis Rheumatol. 2016, 68, 2263–2273. [Google Scholar] [CrossRef] [PubMed]
- Escámez, M.J.; García, M.; Larcher, F.; Meana, A.; Muñoz, E.; Jorcano, J.L.; Del Río, M. An in vivo model of wound healing in genetically modified skin-humanized mice. J. Investig. Dermatol. 2004, 123, 1182–1191. [Google Scholar] [CrossRef] [Green Version]
- Escámez, M.J.; Carretero, M.; García, M.; Martínez-Santamaría, L.; Mirones, I.; Duarte, B.; Holguín, A.; García, E.; García, V.; Meana, A.; et al. Assessment of optimal virus-mediated growth factor gene delivery for human cutaneous wound healing enhancement. J. Investig. Dermatol. 2008, 128, 1565–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Santamaría, L.; Conti, C.J.; Llames, S.; García, E.; Retamosa, L.; Holguín, A.; Illera, N.; Duarte, B.; Camblor, L.; Llaneza, J.M.; et al. The regenerative potential of fibroblasts in a new diabetes-induced delayed humanised wound healing model. Exp. Dermatol. 2013, 22, 195–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escamez, M.; Carretero, M.; García, M.; Martínez-Santamaría, L.; Meana, A.; Larcher, F.; del Rio, M. Smart Growth Factor Gene Delivery for Impaired Wound Healing. In Advances in Wound Care: Volume 1; Sen, C.K., Ed.; Mary Ann Liebert Inc.: New Rochelle, NY, USA, 2010; pp. 367–374. [Google Scholar] [CrossRef]
- Broadbent, J.; Walsh, T.; Upton, Z. Proteomics in chronic wound research: Potentials in healing and health. Proteom. Clin. Appl. 2010, 4, 204–214. [Google Scholar] [CrossRef] [Green Version]
- Nuutila, K.; Siltanen, A.; Peura, M.; Bizik, J.; Kaartinen, I.; Kuokkanen, H.; Nieminen, T.; Harjula, A.; Aarnio, P.; Vuola, J.; et al. Human skin transcriptome during superficial cutaneous wound healing. Wound Repair Regen. 2012, 20, 830–839. [Google Scholar] [CrossRef]
- Pastar, I.; Wong, L.L.; Egger, A.N.; Tomic-Canic, M. Descriptive vs mechanistic scientific approach to study wound healing and its inhibition: Is there a value of translational research involving human subjects? Exp. Dermatol. 2018, 27, 551–562. [Google Scholar] [CrossRef] [Green Version]
- Hidalgo, M.R.; Cubuk, C.; Amadoz, A.; Salavert, F.; Carbonell-Caballero, J.; Dopazo, J. High throughput estimation of functional cell activities reveals disease mechanisms and predicts relevant clinical outcomes. Oncotarget 2017, 8, 5160–5178. [Google Scholar] [CrossRef] [Green Version]
- Çubuk, C.; Hidalgo, M.R.; Amadoz, A.; Rian, K.; Salavert, F.; Pujana, M.A.; Mateo, F.; Herranz, C.; Carbonell-Caballero, J.; Dopazo, J. Differential metabolic activity and discovery of therapeutic targets using summarized metabolic pathway models. NPJ Syst. Biol. Appl. 2019, 5, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meana, A.; Iglesias, J.; Del Rio, M.; Larcher, F.; Madrigal, B.; Fresno, M.F.; Martin, C.; San Roman, F.; Tevar, F. Large surface of cultured human epithelium obtained on a dermal matrix based on live fibroblast-containing fibrin gels. Burns 1998, 24, 621–630. [Google Scholar] [CrossRef]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Al-Shahrour, F.; Minguez, P.; Vaquerizas, J.M.; Conde, L.; Dopazo, J. BABELOMICS: A suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 2005, 33, W460–W464. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Hameedaldeen, A.; Liu, J.; Batres, A.; Graves, G.S.; Graves, D.T. FOXO1, TGF-β regulation and wound healing. Int. J. Mol. Sci. 2014, 15, 16257–16269. [Google Scholar] [CrossRef] [Green Version]
- Liarte, S.; Bernabé-García, Á.; Nicolás, F.J. Role of TGF-β in Skin Chronic Wounds: A Keratinocyte Perspective. Cells 2020, 9, 306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salgado, G.; Ng, Y.Z.; Koh, L.F.; Goh, C.S.M.; Common, J.E. Human reconstructed skin xenografts on mice to model skin physiology. Differentiation 2017, 98, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Arbieva, Z.H.; Guo, S.; Marucha, P.T.; Mustoe, T.A.; DiPietro, L.A. Positional differences in the wound transcriptome of skin and oral mucosa. BMC Genom. 2010, 11, 471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- St Laurent, G., 3rd; Seilheimer, B.; Tackett, M.; Zhou, J.; Shtokalo, D.; Vyatkin, Y.; Ri, M.; Toma, I.; Jones, D.; McCaffrey, T.A. Deep Sequencing Transcriptome Analysis of Murine Wound Healing: Effects of a Multicomponent, Multitarget Natural Product Therapy-Tr14. Front. Mol. Biosci. 2017, 4, 57. [Google Scholar] [CrossRef]
- Wu, C.; Chen, X.; Shu, J.; Lee, C.T. Whole-genome expression analyses of type 2 diabetes in human skin reveal altered immune function and burden of infection. Oncotarget 2017, 8, 34601–34609. [Google Scholar] [CrossRef] [Green Version]
- Takematsu, E.; Spencer, A.; Auster, J.; Chen, P.C.; Graham, A.; Martin, P.; Baker, A.B. Genome wide analysis of gene expression changes in skin from patients with type 2 diabetes. PLoS ONE 2020, 15, e0225267. [Google Scholar] [CrossRef] [Green Version]
- Ferroni, L.; Gardin, C.; Dalla Paola, L.; Campo, G.; Cimaglia, P.; Bellin, G.; Pinton, P.; Zavan, B. Characterization of Dermal Stem Cells of Diabetic Patients. Cells 2019, 8, 729. [Google Scholar] [CrossRef] [Green Version]
- Jere, S.W.; Houreld, N.N.; Abrahamse, H. Role of the PI3K/AKT (mTOR and GSK3β) signalling pathway and photobiomodulation in diabetic wound healing. Cytokine Growth Factor Rev. 2019, 50, 52–59. [Google Scholar] [CrossRef]
- Loots, M.A.; Lamme, E.N.; Zeegelaar, J.; Mekkes, J.R.; Bos, J.D.; Middelkoop, E. Differences in cellular infiltrate and extracellular matrix of chronic diabetic and venous ulcers versus acute wounds. J. Investig. Dermatol. 1998, 111, 850–857. [Google Scholar] [CrossRef] [Green Version]
- Shaw, T.J.; Martin, P. Wound repair at a glance. J. Cell Sci. 2009, 122, 3209–3213. [Google Scholar] [CrossRef] [Green Version]
- Peake, M.A.; Caley, M.; Giles, P.J.; Wall, I.; Enoch, S.; Davies, L.C.; Kipling, D.; Thomas, D.W.; Stephens, P. Identification of a transcriptional signature for the wound healing continuum. Wound Repair Regen. 2014, 22, 399–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Ma, K.; Kwon, S.H.; Garg, R.; Patta, Y.R.; Fujiwara, T.; Gurtner, G.C. The Abnormal Architecture of Healed Diabetic Ulcers Is the Result of FAK Degradation by Calpain 1. J. Investig. Dermatol. 2017, 137, 1155–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatanaka, E.; Monteagudo, P.T.; Marrocos, M.S.; Campa, A. Neutrophils and monocytes as potentially important sources of proinflammatory cytokines in diabetes. Clin. Exp. Immunol. 2006, 146, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Arya, A.K.; Tripathi, R.; Kumar, S.; Tripathi, K. Recent advances on the association of apoptosis in chronic non healing diabetic wound. World J. Diabetes 2014, 5, 756–762. [Google Scholar] [CrossRef]
- Brem, H.; Stojadinovic, O.; Diegelmann, R.F.; Entero, H.; Lee, B.; Pastar, I.; Golinko, M.; Rosenberg, H.; Tomic-Canic, M. Molecular markers in patients with chronic wounds to guide surgical debridement. Mol. Med. 2007, 13, 30–39. [Google Scholar] [CrossRef]
- Sawaya, A.P.; Jozic, I.; Stone, R.C.; Pastar, I.; Egger, A.N.; Stojadinovic, O.; Glinos, G.D.; Kirsner, R.S.; Tomic-Canic, M. Mevastatin promotes healing by targeting caveolin-1 to restore EGFR signaling. JCI Insight 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Ponugoti, B.; Tian, C.; Xu, F.; Tarapore, R.; Batres, A.; Alsadun, S.; Lim, J.; Dong, G.; Graves, D.T. FOXO1 differentially regulates both normal and diabetic wound healing. J. Cell Biol. 2015, 209, 289–303. [Google Scholar] [CrossRef] [Green Version]





| Comparison | Differentially Expressed Probes | Up-Regulated Probes | Down-Regulated Probes | Significant GO_BPs | Significant KEGG |
|---|---|---|---|---|---|
| C24 vs. C0 | 7570 | 3350 | 4220 | 456 | 42 |
| D0 vs. C0 | 403 | 101 | 302 | 100 | 7 |
| D24 vs. C24 | 49 | 14 | 35 | 3 | 0 |
| D24 vs. D0 | 8686 | 3801 | 4885 | 519 | 71 |
| Comparison | Significant GO_BPs | Significant KEGG | ||
|---|---|---|---|---|
| Up-Regulated Probes | Down-Regulated Probes | Up-Regulated Probes | Down-Regulated Probes | |
| Unique C24vsC0 | 67 | 122 | 11 | 27 |
| Unique D24vsD0 | 204 | 86 | 72 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
León, C.; García-García, F.; Llames, S.; García-Pérez, E.; Carretero, M.; Arriba, M.d.C.; Dopazo, J.; del Río, M.; Escámez, M.J.; Martínez-Santamaría, L. Transcriptomic Analysis of a Diabetic Skin-Humanized Mouse Model Dissects Molecular Pathways Underlying the Delayed Wound Healing Response. Genes 2021, 12, 47. https://doi.org/10.3390/genes12010047
León C, García-García F, Llames S, García-Pérez E, Carretero M, Arriba MdC, Dopazo J, del Río M, Escámez MJ, Martínez-Santamaría L. Transcriptomic Analysis of a Diabetic Skin-Humanized Mouse Model Dissects Molecular Pathways Underlying the Delayed Wound Healing Response. Genes. 2021; 12(1):47. https://doi.org/10.3390/genes12010047
Chicago/Turabian StyleLeón, Carlos, Francisco García-García, Sara Llames, Eva García-Pérez, Marta Carretero, María del Carmen Arriba, Joaquín Dopazo, Marcela del Río, María José Escámez, and Lucía Martínez-Santamaría. 2021. "Transcriptomic Analysis of a Diabetic Skin-Humanized Mouse Model Dissects Molecular Pathways Underlying the Delayed Wound Healing Response" Genes 12, no. 1: 47. https://doi.org/10.3390/genes12010047
APA StyleLeón, C., García-García, F., Llames, S., García-Pérez, E., Carretero, M., Arriba, M. d. C., Dopazo, J., del Río, M., Escámez, M. J., & Martínez-Santamaría, L. (2021). Transcriptomic Analysis of a Diabetic Skin-Humanized Mouse Model Dissects Molecular Pathways Underlying the Delayed Wound Healing Response. Genes, 12(1), 47. https://doi.org/10.3390/genes12010047