1. Introduction
Antimicrobial resistance in bacteria is not a new phenomenon. Estimates for the emergence of biosynthetic pathways have been published that show this occurring hundreds of millions of years ago [
1]. It has further been documented that bacteria have a pool of shared antimicrobial resistance genes (AMRGs) present in the antibiotic resistome prior to humans’ discovery and widespread use of antibiotics. In permafrost from 30,000 years ago, genes encoding resistance for β-lactam and other antibiotics have been documented [
2]. There are self-transmissible plasmids carrying antibiotic resistance genes (ARGs) that can infect many phylogenetically distinct bacteria creating a pool of genes to share [
3]. However, the role that human actions play in adding to the antibiotic resistome, and spurring a rapid increase of the numbers and diversity of antibiotic resistant bacteria (ARB), is relatively new ramping up with the discovery and wide-spread use of penicillin in the 1940′s. The number of daily antibiotic doses increased from 21.4 to 34.8 between 2000 to 2015, with a 40% daily dose rate increase per thousand inhabitants in 76 countries [
4]. So, while antibiotic resistance is ancient, human activities have increased the numbers and types of antibiotic resistance strategies deployed by bacteria by loading the environment with antibiotics, creating conditions that apply pressure to clinical and environmental bacteria to adapt.
Human usage of antibiotics has resulted in the enhanced resistance of well-known, and opportunistic bacterial pathogens, to multiple antibiotics through mutation and acquisition of ARGs [
1]. About 65% of these antibiotics are β lactam types and their usage continues to increase [
5] Inputs from systems that grow livestock and treat sewage have made wastes generated from these activities the largest, continuous input of antimicrobials and antibiotics into aquatic systems, including the sediments and biofilms within [
6,
7]. With aquatic systems that serve as sinks for antibiotics and resistant organisms generated by human activities, the potential for gene transmission from human and animal pathogens, to environmental bacteria, then back to human and animal pathogens is probable [
8,
9]. Bacteria exchange genetic information horizontally through direct uptake of DNA (transformation), virally-mediated transduction, and pili-mediated conjugation. Sub-lethal levels of antibiotics and other antimicrobials, such as those found in sewage-associated aquatic systems, can stimulate horizontal gene transfer (HGT) and modulate gene transcription in bacteria, creating a flow of resistance genes between environmental and clinically significant bacteria [
8,
9]. The predominant direction of the flow is not well understood, but with other factors creating low fitness costs for the carriage of antimicrobial resistance genes (AMRGs), and documented transmission of these genes to environmental bacteria, aquatic systems are thought to be important, long-term reservoirs for antimicrobial resistance genes. To control the spread of AMRGs, the ways clinical and environmental bacteria share genetic materials must be understood.
ARGs often co-occur with biocidal/metals resistance genes (BMRGs) in mobile genetic elements [
1] and are found in bacteria that are associated with human activity. A recent study of fully sequenced bacterial genomes and plasmids (
n = 4582) has provided more insight into the co-occurrence of ARGs and BMRGs. In situations where the ARGs and BMRGs were located on the same plasmid, the plasmid size was larger (76 kb average) in comparison with plasmids that carried only ARGs [
10]. The larger size of these plasmids was linked with the plasmids being conjugative, which would require close contact between the bacteria for HGT, while the larger size would incur survival costs to the bacteria to retain the plasmid. However, survival costs are lowered by increases in the levels of antimicrobials or metals in the aquatic matrix, and sewage has enhanced levels of both in comparison with natural aquatic systems. Sewage has antibiotics, disinfectants, cleaning compounds, metals, and other antimicrobials in solution, although in sublethal concentrations 100 times lower than that found in hospital sewage [
6]. These concentrations are thought to be linked to an increase in AMRGs, ARGs, and BMRGs in the effluent bacteria that survive disinfection. It has been shown that while enterococci decreased in concentration by log factors through sewage treatment, these bacteria, 50% of which were antibiotic resistant, were still present in up to the thousands of CFU per mL of effluent [
7]. A study reported that the final effluent of an urban water treatment plant could discharge 10
7 to 10
10 antibiotic resistant bacteria per inhabitant [
7]. Clearly sewage effluent is a source of antibiotic resistance into our aquatic systems.
Staphylococci bacteria have been found in sewage and sewage effluents. Staphylococci are often isolated with several other species co-growing as skin commensals. Many species can cause infections of the skin or blood and organs, and a quick way to differentiate the members of this family is by coagulase testing. There are numerous types of coagulase-positive staphylococci (CoPS), from the well-known pathogenic, methicillin resistant
Staphylococcus aureus (MRSA) of clinical significance to humans that has also been found in livestock, to
S. delphini, a strain of veterinary concern that has recently had a documented human case reported [
11]. There are clinically significant, coagulase-negative staphylococci (CoNS) as well:
S. epidermidis and
S. warneri, both commensal bacteria that can opportunistically cause device-associated infections, although more rarely for
S. warneri. Of the CoNS skin commensals,
S. epidermidis is isolated in greater numbers and more frequently from the skin than
S. warneri [
12], which comprises <1% of the total Staphylococcal skin flora [
13]. Staphylococci have numerous mobile genetic elements, and readily exchange genetic materials through HGT. There is high homology between CoNS and CoPS
mecA cassettes [
14], and the cassette has been isolated from several species of staphylococci. Of the plasmids investigated for
S. aureus, 70% had AMRGs with 20% carrying both ARGs and BMRGs [
10].
The function of antibiotics and AMRGs in clinical settings changes after releasing them into the non-clinical environment [
15]. Wastewater treatment plants are counted as a significant, anthropogenic source for spreading antibiotics, AMRGs, and antibiotic resistance bacteria by releasing their effluents into the natural waters [
7]. The presence of
S. aureus with the carriage of ARGs and MRGs has been reported in wastewater in multiple studies [
16,
17,
18,
19]. However, the possible mobile genetic elements (plasmids, transposons, bacteriophages, and integrons) for hosting resistance genes and potential pathways for exchanging, acquiring, and spreading them are vague among environmental staphylococci species [
7]. Metagenomic studies have shown the presence of different resistance genes on sewage plasmids, and the significant role of these plasmids in transferring resistance genes through HGT [
20]. It is necessary to identify the potential conditions that facilitate the acquisition of resistance genes of
S. aureus origins to control their spread and retard future evolutions. Historically, plasmids were classified based on the type of replication (Rep) protein [
21]; also named replicon [
22]. Different selective pressures affect the diversity of plasmids. This means that the presence of a plasmid which encodes ARGs is relative to the clinical origins of the parent strain [
23].
In this study, the prevalence and co-carriage of ARGs and MRGs were investigated in sewage-associated staphylococci species. Whole genome sequencing and BLASTN analysis was coupled to phenotypic screenings to detect plasmid-encoded resistance genes and determine bacterial origin of these mobile genetic elements in CoNS and CoPS. The growth of the more numerous S. epidermidis was suppressed to focus on potential gene sharing between identified strains of S. aureus, S. warneri, and S. delphini from isolates collected from sewage and sewage-impacted aquatic environments. Our prior research had shown the carriage of MRGs thought to originate in S. aureus, in S. warneri, and vice versa (results and data under review). It was also noted that S. warneri isolates were carrying MRGs originating from S. delphini. To further this work, we analyzed the ARGs in these isolates, and investigated if the ARGs (such as the blaZ gene) and MRGs (cadD and cadX genes) were carried in the sequences of identifiable plasmids, the plasmid partial or complete presence, and if the blaZ, cadD, and cadX were carried on plasmids in S. aureus and S. warneri or were present without being located in the identifiable plasmids.
3. Results
3.1. Types of Isolates Based on Whole Genomic Sequencing
In this study, 24 sequenced sewage-associated staphylococci isolates were selected from all isolates archived to cover different sampling locations and different days. These 24 isolates were sequenced to investigate if MRGs and ARGs were present, and if they were located within plasmids. These isolates were obtained from sewage (influent, effluent, and sedimentation tank), or from a creek impacted by a wastewater treatment plant effluent (sediments from West Hickman creek), or where leaking sewers impacted sediment samples in a local park creek (Veterans Park creek-sediment). Among the 24 staphylococci isolates, twenty of them were identified as
S. aureus, including 17 isolates found in wastewater (ID:1–18) and three isolates detected from creek sediment samples (ID: 51, 52, and 60). Two isolates were
S. warneri (ID 54 and 55) isolated from the final chlorinated effluent, and two
S. delphini isolates (ID: 53 and 56) were found in upstream sediment samples impacted by sewage leaks (ID:53), or downstream sediments impacted by the effluent of WWTP (ID:54) with one month differences in collection times. Classification of the
S. aureus isolates showed the presence of three clonal complexes, CC59, CC5, CC8 with a number of different ARGs (
Table 1).
Isolate #2 classified as CC59, which was the only isolate identified as the prevalent clonal complex in community isolates identified in Asiatic countries such as China, Taiwan, and Vietnam. It was recovered from a sewershed that had an airport, several hospitals, several college campuses, and high population density. Among the other of 19 S. aureus isolates, six of them typed as hospital-related S. aureus (CC5), and 13 isolates were classified as an environmentally-circulating strain of S. aureus (CC8). In this study all six isolates in CC5 subtyped as ST5. Isolates in CC8 had four different STs including ST8 (the ancestral genotype), ST2642 and ST1750 (single locus variants of ST8), and ST72 (triple locus variant).
3.2. Types of ARGs in Sewage-Associated Staphylococcus Isolates
Among the staphylococcus species in this study, five groups of antibiotic resistance phenotypes were detected as shown in
Table 1 including β lactam, macrolide, streptogramin, aminoglycoside, and chloramphenicol antibiotics. As shown in
Table 1, the
blaZ gene was found in all
S. aureus isolates except for three CC5-ST5 isolates (#13, 15, 18). All CC8 isolates carried the
blaZ gene, but only 50% of CC5 isolates carried
blaZ gene. The
mecA gene was not as prevalent as the
blaZ gene for β lactam resistance, with only 16% of CC5 (only 1 of 6 isolates) and 76% of CC8 (3 of 13) isolates positive. The
mecA positive isolates were classified as methicillin-resistant
S. aureus (MRSA) based on
mecA presence. Based on these observations, it seems that the
blaZ gene is well conserved in sewage-associated
S. aureus and often associated with
mecA in CC8 isolates. The presence of the
blaZ gene may be related to the host type, and it is notable that both β lactam ARGs were absent in
S. warneri and
S. delphini isolates, which are animal associated.
For
S. aureus CC8 isolates, only once was carriage of ARGs limited to a single gene (#8); and for those that had two ARGs, the second gene was most often
mecA. Comparing the number of ARGs between different isolate types in CC8 shows that some strains carried more ARGs than others, with multiple types of antibiotic resistance common. In general, the average number of ARGs per
S. aureus clonal complex, and the types of antibiotic class resistance, was higher in CC8-ST1750 (single locus variant of ST8) than for other
S. aureus isolates, with isolate #3 carrying the maximum of six ARGs covering β lactam, macrolide, aminoglycoside, and chloramphenicol antibiotics. A second group of
S. aureus isolates with higher numbers of ARGs was CC8-ST72, which is a triple locus variant of ST8 and frequently isolated in China and Korea and often harbouring numerous toxins [
29]. Both isolate groups came from antibiotic-rich environments (WWTP influent or creek sediment under the influence of leaking sewers). It appears that within types of a clonal complexes, slight variations were associated with increased ARG carriage.
Isolates with only a single antibiotic-resistance gene were more prevalent in CC5 group (50%), and the resistance for macrolides more prevalent than for other antibiotic groups with, 66% of isolates positive for the presence of erm(A), in contrast with 23% for all CC8 isolates. The other species of staphylococci had less ARGs present than S. aureus. Both S. warneri isolates (ID#: 54, 55) from fully treated WWTP effluent did not carry any β lactam antibiotic-resistance genes, but both isolates carried a unique type of antibiotic-resistance gene (Vga(A)LC) belonging to streptogramin phenotype, and one had an additional erm(C) resistance gene for macrolides. The Vga(A)LC gene was not found in the sequence of any other sewage-associated staphylococci isolates. The two S. delphini isolates from creek sediments were negative for the carriage of any ARGs.
Thus far, multiple research groups have reported the carriage of the
blaZ, cadmium, and arsenic resistance genes on plasmids [
30]. Therefore, the concurrence of these resistance genes was further investigated to illuminate possible associations between MRGs, ARGs, and plasmid transference in sewage-associated staphylococci isolates.
3.3. Types of Plasmids in Isolates
Table 2 summarizes the list of identified plasmids in this study with their accession numbers, lengths, rep numbers, and the specific bacterial types.
As shown in
Table 2, the shortest plasmid in
S. aureus group is the rep 10 plasmid (Accession #: GU562624) with 2402 base pairs, while the rest of plasmids in this group have lengths between 24,653 to 57,889. As shown in
Table 2, plasmids were classified into three groups according to labelled species origination for reference plasmids specific to
S. aureus,
S. warneri, and
S. epidermidis. The presence of selected plasmids, and the percent of the plasmid found in different isolates sequences are summarized in
Table 3.
What is very clear is that S. aureus isolates carried a number of plasmids, but each clonal complex had differences in carriage, type, and number. The sewage-associated S. aureus isolates carried one to four plasmids, except for two isolates from CC5-ST5, (#13 from influent and #18 from effluent) that did not carry any plasmids, although they had shown carriage of erm(A) ARG. In general, CC8 isolates carried only single S. aureus plasmids, while CC5 carried multiple plasmids for 3 of the 4 plasmid carrying isolates.
There were some unique plasmid carriages associated with staphylococci species and
S. aureus clonal complexes. Of the ten plasmids listed in
Table 3, five of the plasmids originated, and were carried only in, a single species, or clonal complex of a species. The unique carriage of plasmids originating within a clonal complex of
S. aureus, or a staphylococcus species, is indicative of the ability of these plasmids to be conserved within a group, with higher degrees of coverage indicating more recent acquisition and greater conservation. It is also suggestive of vertical gene transmission.
The rep 13 and 15 plasmids were found only in S. aureus CC5 isolates. The single S. aureus CC59 isolate (#2), carried a unique plasmid, the rep 5a, which was not found in any other isolates. The whole length of the rep5a plasmid was encoded in isolate #2 (100%). Both S. warneri isolates uniquely carried a plasmid labelled as originating in S. warneri (rep13a-AB125342) with 88% coverage as compared to the reference sequence.
Within the S. aureus isolates there were plasmids whose presence was not confined within a single clonal complex. The rep 19 plasmid was carried in one CC5 isolate (#14), and seven CC8 isolates, including four isolates of ST8 with only partial (52%) of sequence coverage, and three isolates of ST1750 with 52% and less coverage. The alignment results showed the exact same sequence of the partial 52% coverage of the rep 19 plasmid (8727 to 21,618 nucleotides) was encoded in all 4 isolates of ST8 and the single ST1750 sediment isolate (#51). Another shared within S. aureus species plasmid was the rep 20 (GQ900426). The rep 20 (GQ900426) plasmid was found in 50% of CC5 isolates and 23% of CC8 isolates, with varying degrees of coverage from 38% to 100%.
There were plasmids that appear to be shared between staphylococci species. S. aureus isolate #7 carried a S. aureus specific rep 10 plasmid at 100% coverage that was also found in one effluent S. warneri isolate sequence (#54). This S. warneri isolate (#54) contained 98% coverage of the sequence of the rep 10 plasmid in its genome. This was the only time that a plasmid (the rep10) was found both in the originating species, and a different staphylococci species in the isolates analysed. Some S. aureus isolates carried plasmids specific to other staphylococci species (S. warneri and S. epidermidis), but these plasmids were not present in the other species isolates that had been sequenced. In addition to the carriage of plasmids specific to S. aureus, isolate #1 (CC8-ST8) carried the rep13 plasmid specific to S. warneri with 42% query coverage, and over 99% identity, which means that 42% of this plasmid (E-value = 0) from another species was encoded with great accuracy in the sequence of isolate #1. In addition, S. aureus isolates #11 and #10 carried a partial sequence of the rep 20 plasmid specific to S. epidermidis with maximum carriage of 34% and 15%, respectively. Unlike isolates in the CC8 group, which captured some parts of plasmid’s sequences related to S. warneri and S. epidermidis, isolates belonging to CC5 group did not have any plasmids specific to coagulase-negative staphylococcus species.
Both S. warneri isolates (ID #54 and #55) isolated from final chlorinated effluent samples carried one plasmid specific to the coagulase-negative species S. epidermidis, the rep 5b plasmid with 58% query coverage. For both isolates the same part of the plasmid, 1956 to 5577 nucleotides, were encoded within the plasmid sequence and the sequence of two isolates were similar with E values equal to zero. While the presence of this partial plasmid is suggestive of horizontal gene transmission, the two isolates were found in effluent only a few days apart, and vertical gene transmission that retained the partial plasmid in the S. warneri that were able to persist in the treatment plant is also suspected.
The two, coagulase-positive S. delphini isolates were the only other type of CoPS species isolated in this study, and both were found in sediment samples from a sewage impacted creek at locations upstream and downstream of the WWTP with one-month difference in sampling. However, neither isolate carried any identifiable plasmids despite having three MRGs specific to S. delphini including csoR (copper-resistant), arsB and arsC (arsenic resistance) (data under review).
After identifying the unique and shared plasmids in each isolate, more analysis was conducted to try and link the presence of MRGs and ARGs to carriage of these plasmids.
3.4. Carriage of MRGs and ARGs on Plasmids
The results for the carriage of MRGs and ARGs on identified plasmids is summarized in
Table 4. The two
S. delphini isolates were not included in this table since they did not have any plasmids identified on their genome, although as mentioned they carried multiple MRGs specific to
S. delphini (results did not present here).
The
blaZ gene was the most prevalent ARG in the
S. aureus isolates, but was not always carried within a plasmid. As shown in
Table 4, 85% (17 out of 20) of the
S. aureus isolates were positive for carrying the
blaZ gene in their sequence, with 100% of the CC8 isolates positive. Among these
blaZ positive isolates, 65% (11 out of 17) contained the
blaZ gene within a plasmid. All 3 genes (
blaZ,
cadD, and
cadX) co-occurred 13 times (two last columns in
Table 4), with co-location on a plasmid detected 8 times (62%).
S. aureus isolates were also carrying the
S. aureus reference
cadD and
cadX MRGs within the same plasmid, but not co-located with
blaZ. Three
S. aureus isolates (#9, #12, and #3) with the
blaZ gene did not carry any MRGs on their plasmid. The carriage of the
blaZ,
cadD, and
cadX within the same plasmid was more common in CC8 isolates (5 out of 13 isolates, 38%) in comparison with CC5 (1 out of 6 isolates, 16%). The order of these ARGs and MRGs carriage was not consistent in all isolates. As shown in this table, in some isolates (#7, #6, #16, and #51) the
blaZ gene was encoded after the cadmium resistance genes. However, in isolates #11 and #5, the
blaZ gene was encoded before cadmium resistance genes.
The unique plasmid, rep 5a, in the single
S. aureus CC59 isolate #2 encoded arsenic and cadmium resistance genes with the
blaZ gene within its sequence. The antibiotic resistant gene
erm(C) in isolate #7 was encoded on the rep 10 plasmid, not with the cadmium resistance genes. This finding supports previous research that has related the association of the
erm(C) to the rep 10 plasmid [
31].
Both blaZ-negative S. warneri isolates, ID# 54 and 55, isolated from the effluent of a WWTP were positive for 3 and 2 different plasmids, respectively. However, the metal (S. aureus cadD and cadX) and antibiotic resistance (erm(C) and Vga(A)LC) genes were not carried within the identified plasmid sequences. This is indicative of the acquisition of ARGs and MRGs specific to S. aureus, and carried on S. aureus plasmids, by S. warneri isolates by other means than directly exchanging S. aureus specific plasmids by conjugation.
4. Discussion
A plasmid analysis of sewage-associated staphylococcus species was conducted in this study to investigate if MRGs and ARGs encoded on plasmids were carried by more than one species, or clonally related to a species. In addition, the co-location of ARGs and MRGs on plasmids was investigated to see if HGT between species, and strains of species, was likely by conjugation. Understanding the relationship between the reference plasmids and encoded MRGs and ARGs is important to comprehend how these resistance genes may spread in the sewage-associated environment. We showed that the presence of different plasmids is related to the lineage of
S. aureus since rep19 and 20 were found in most of isolates in CC8 and CC5 groups. These two clonal complexes are widely distributed in human and veterinary medicines. Different plasmid groups of
S. aureus isolates were mentioned by McCarthy and Lindsay (28) to show what MRGs, ARGs, and virulent genes are encoded on different plasmids in each group. Based on their findings, the
blaZ and the
cadDX genes are two variable resistance genes (they are not core genes but transferred by mobile genetic elements) that occur together on the rep 19 plasmid [
31]. The linked carriage of the
blaZ and cadmium resistance genes (
cadD and
cadX) was shown in another study [
32] and our results support both of these studies findings as our observations also related the encoding of the
cadD,
cadX and
blaZ genes together on the same plasmid. These markers are quite abundant in strains isolated from environmental samples and are not restricted to Gram-positive bacteria but are also present in several Gram-negative species [
33,
34,
35].
The McCarthy and Lindsay (2012) study documented the presence of rep 5 in
S. aureus CC15, CC25, CC30, and CC45 [
31]. Our findings expand theirs by documenting the rep 5 plasmid in CC59, a group of community
S. aureus strains prevalent in numerous Asian countries. This difference in our findings could be related to the different environment (country-origin) that their isolates came from, or to a more recent acquisition of rep 5a by the CC59 isolate in our study. Also, as they noted the presence of the
blaZ gene in all human lineages of
S. aureus was documented, yet was rare among animal-associated
S. aureus [
31]. In this study, all
S. aureus isolates except three CC5-ST5 isolates were positive for the presence of the
blaZ gene. This is suggestive that our sewage-associated
S. aureus isolates came predominately from human hosts where co-growth in skin niches could encourage HGT by conjugation, while stressors in this environment could encourage vertical transmission after gene acquisition.
The co-carriage of blaZ and cadmium MRGs commonly coded within S. aureus plasmids can be related to the environmental conditions around the bacteria that impact gene transfer. These encoded resistance genes on plasmids are exchanged via HGT with other S. aureus and staphylococci bacteria. The exchange of large, complete plasmids between two bacteria by conjugation are favoured in high-density conditions when bacteria have a direct contact to each other. That plasmid conjugation occurred can be inferred from our study results where a number of isolates contained large plasmids (rep 19 and 20) present in S. aureus at good levels of coverage, and had the blaZ, cadD, and cadX resistance genes located within those plasmids. However, the presence of these plasmids, even at greater coverages, did not assure the co-carriage of blaZ and cadmium resistance genes within the plasmid. The presence of rep 20 plasmids at 100% coverage in S. aureus CC8- ST2642 isolates #9 and #12 did not have the MRGs encoded within the plasmid, even though these genes were present in the isolate. So, the presence of the rep 19 and 20 plasmid was not always associated with the presence of the blaZ and cadmium resistance genes within the plasmid.
In this study, 35% (7 out of 20) of S. aureus isolates carried the blaZ gene with cadD and cadX gene on the same plasmid, one isolate carrying these genes in two separate plasmids. The plasmid most frequently carrying all three genes was the rep 19, which occurred 6 of the 8 times. If we consider all S. aureus isolates that carried ARGs, the co-carriage of both cadmium resistance genes and ARGs in a plasmid sequence occurred in 40% (8 of 20) of the S. aureus isolates in this study. This percentage of co-occurrence of ARGs and MRGs is higher than reported by Pal et al. (9) for 4582 plasmid sequences from publicly available bacterial genomes, only 5% of plasmids carried both ARGs and BMRGs. They documented 20% of co-occurrence of ARGs and BMRGs on plasmids for Staphylococcus genomes (9), which is less than half the frequency we found for our isolates, even though we selected for only cadmium-specific MRGs instead of any AMRGs co-occurrence with ARGs. It would seem that there is more research needed on the co-carriage on ARGs and AMRGs in sewage-associated environments to help understand the changes in the resistome present in polluted aquatic systems.
For other sewage-associated staphylococci species, the lack of plasmids and presence of ARGs and MRGs outside of plasmids, suggests that the acquisition of genes from sewage-associated
S. aureus can occur by other gene transfer mechanisms. As mentioned by Pal et al. [
10], the frequency of co-carriage of ARGs and BMRGs for the bacteria they examined was greater in larger plasmids, with a median size of 76 kb. In our study, the size of plasmids (rep 19, 20, and 5a) carrying
blaZ,
cadD, and
cadX were smaller than 30.4 kb, and even smaller when taking into consideration that often only partial plasmids were found (<53% coverage rep 19). Their study did show a number of ARGS and AMRGs in plasmids of 25kb average size, but none in plasmids less than 20 kb (9), suggesting that the mode of gene transfer for these smaller, partial plasmids, was not conjugative.
For the two
S. warneri isolates, conjugation of plasmids does not appear to be the primary mode for acquisition of
S. aureus reference cadmium MRGs, nor macrolide and streptogramin B ARG acquisition since neither of these isolates had any plasmids identified that carried the genes. This lack of plasmid co-carriage of ARGs and MRGs could be related to its hosts and prevalence in the broader environment.
S. warneri are known to inhabit animal hosts [
36] as well as humans, and have been implicated in human disease [
37]. However, even though
S. warneri may grow in co-culture on animal hosts in small numbers, in sewage-associated aquatic systems, we did not find co-carriage of MRGs and ARGs on plasmids, suggesting another mechanism for HGT acquisition of these genes from other species of bacteria.
S. warneri can co-grow on human hosts as it has been reported to be the next most frequently identified, coagulase-negative, methicillin resistant, staphylococcus species after
S. epidermidis found colonizing baby-care personnel and is known to carry the SCC cassette that encodes for
mecA [
38,
39,
40]. However,
S. warneri is not as commonly found on humans as other staphylococci, often found on animals, and our study found a lesser prevalence in untreated sewage than
S. aureus. Only in the treated sewage effluent, after chlorination, was
S. warneri the dominant species consistently isolated from samples.
S. aureus was isolated by selective growth enrichment from effluent only once in two years of sampling by comparison. It is unknown if S.
warneri can grow inside the sewage treatment plant, but the continuous presence in the treatment plant effluent after chlorination points to an ability to survive, and perhaps even grow in these conditions.
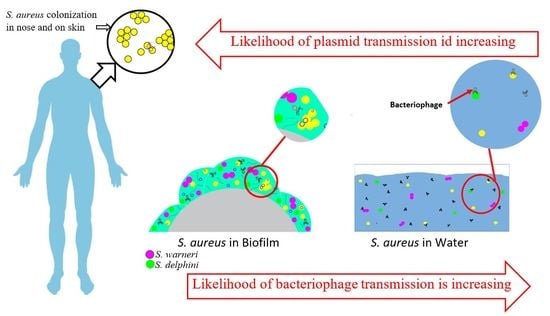
In the effluent environment, after sedimentation has cleared microbially dense sludge biofilms from the water column resulting in lower bacterial densities, genetic information can still be transferred by transduction, using unsettleable bacteriophage as vectors between bacteria, or by transformation. Generalized transduction could result in the uptake of pieces of genetic material from other bacteria in the DNA and bacteriophage-loaded effluent, leading to the acquisition of genes without the acquisition of complete plasmids, especially in the presence of different environmental stresses such as toxic compounds or biocidal molecules [
38]. As mentioned by Pal et al. [
10], HGT of ARGs is increased by exposure to metals and biocides, and they suggested that use of zinc and cadmium supplementation could potentially promote co-selection for, and HGT of, ARGs and BMRGs. In our study results, we found cadmium resistance reference gene sequences for
S. aureus in
S. warneri, which suggests HGT between these staphylococci species. However, the presence of reference
S. aureus cadmium resistance genes and absence of
blaZ, without the presence of the rep 19 and 20 plasmids known to encode these resistance genes together, along with the presence of
erm(C) without the rep 10 plasmid, suggests that the two
S. warneri isolates acquired MRGs and ARGs via HGT by transduction, or direct DNA uptake, from non-viable
S. aureus.
The acquisition of resistance genes from other staphylococci associated with escalating disease burdens, into S. warneri that are being consistently released into the environment from our WWTPs in large numbers, is concerning and potentially hazardous to animals and people encountering ARB in receiving waters. S. warneri has been shown by others to carry mecA and have methicillin resistance. The acquisition of plasmids specific to S. aureus (the rep 10) and S. epidermidis (the rep 5b) provide more confirmation about the ability of S. warneri to obtain, carry, and spread plasmid-associated resistance genes from organisms shed from human hosts into aquatic systems via transport by WWTP effluent into receiving streams. As WWTP effluent can provide a significant source of water in receiving streams, especially in arid regions, the potential for S. warneri to obtain virulence genes and pass them to other bacteria needs to be researched so this risk can be fully understood.
Our results showed how cadmium resistance genes were shared between S. aureus and S. warneri sewage isolates, but did not show that these other staphylococci were capable of sharing genes with S. delphini isolates present in the receiving creek sediments. The environmental conditions, or the resistance of S. delphini, seems to be preventing gene transference, as indicated by the lack of plasmids, ARGs, and MRGs from other staphylococci bacteria in both S. delphini isolates. The analysis of MRGs (data did not presented here) showed that these two S. delphini isolates did not carry any MRGs specific to S. aureus or S. warneri. However, MRGs specific to S. delphini were found in both S. delphini isolates, and other staphylococcus species isolates in this study, suggesting that while HGT between species can occur, it is limited due to differences in the staphylococci species ability to acquire and spread virulence factors from human pathogens to other bacteria. A better understanding of the resistance of S. delphini to HGT may help us lessen the transference in other staphylococci.
ARGs carriage was variable among the
S. aureus isolates by type. Macrolide resistance was detected in a greater percentage in CC5 isolates in this study, which was different from reported results by McCarthy and Lindsey [
31] since the presence of the
erm(A) gene was reported in CC8 and CC239 lineages and not in CC5 in their study. Macrolide resistance was found to be more prevalent in CC8 isolates, but not in the ST8 group of isolates.
Our results showed that isolates from two types of
S. aureus, CC8-ST 1750 (single locus variant of ST8) and CC8-ST72 (triple locus variant of ST8), carried more ARGs from isolates found in wastewater and sediment in comparison with other CC8 types. This could be related to their emergence as human pathogens, with more host carriage, or to the evolution of greater survival in the antibiotic-loaded environment of sewage [
7].