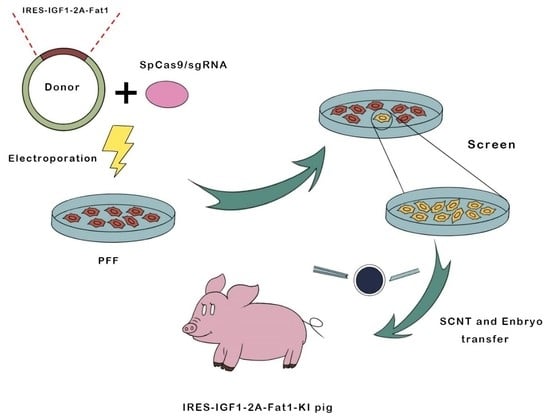
CRISPR/Cas9-Mediated Specific Integration of Fat-1 and IGF-1 at the pRosa26 Locus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Vector Construction
2.2. Electroporation and Limiting Dilution Screening of PFFs
2.3. SCNT and Embryo Transfer
2.4. Generation of Transgenic Pigs and Genotype Analysis
2.5. Transcriptional Analysis of the IGF-1 and Fat-1 Genes in Transgenic Pigs Using Real-Time PCR
2.6. Western Blot Analysis
2.7. Fatty Acid Analysis
2.8. Statistical Methods
3. Results
3.1. Construction of the Fat-1 and IGF-1 Donor Vector and CRISPR/Cas9-Mediated Integration of Fat-1 and IGF-1 into PFFs
3.2. F0 Generation of Knock-in Pigs and Transcriptional Analysis of the Fat-1 and IGF-1 Genes
3.3. Analysis of the IGF-1 Protein and Fatty Acids in the IGF-1 and Fat-1 Knock-in Pigs
3.4. Off-Target Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, C.; Pang, D.; Li, M.; Yuan, H.; Yu, T.; Huang, P.; Li, J.; Chen, X.; Jiao, H.; Xie, Z.; et al. Crispr/cas9-mediated hitchhike expression of functional shrnas at the porcine mir-17-92 cluster. Cells 2019, 8, 113. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Ouyang, H.; Xie, Z.; Yao, C.; Guo, N.; Li, M.; Jiao, H.; Pang, D. Efficient generation of myostatin mutations in pigs using the crispr/cas9 system. Sci. Rep. 2015, 5, 16623. [Google Scholar] [CrossRef]
- Xie, Z.; Pang, D.; Yuan, H.; Jiao, H.; Lu, C.; Wang, K.; Yang, Q.; Li, M.; Chen, X.; Yu, T.; et al. Genetically modified pigs are protected from classical swine fever virus. PLoS Pathog. 2018, 14, e1007193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, H.; Yu, T.; Wang, L.; Yang, L.; Zhang, Y.; Liu, H.; Li, M.; Tang, X.; Liu, Z.; Li, Z.; et al. Efficient base editing by RNA-guided cytidine base editors (cbes) in pigs. Cell. Mol. Life Sci. 2020, 77, 719–733. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Ouyang, H.; Yu, T.; Chen, X.; Pang, D.; Tang, X.; Chen, C. Preparation of a new type 2 diabetic miniature pig model via the crispr/cas9 system. Cell Death Dis. 2019, 10, 823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Jiao, H.; Xiao, H.; Jiang, Y.; Liu, Z.; Qi, C.; Zhao, D.; Jiao, S.; Yu, T.; Tang, X.; et al. Generation of prsad2 gene knock-in pig via crispr/cas9 technology. Antiviral Res. 2020, 174, 104696. [Google Scholar] [CrossRef]
- Pan, J.; Tao, C.; Cao, C.; Zheng, Q.; Lam, S.M.; Shui, G.; Liu, X.; Li, K.; Zhao, J.; Wang, Y. Adipose lipidomics and RNA-seq analysis revealed the enhanced mitochondrial function in ucp1 knock-in pigs. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 1375–1383. [Google Scholar] [CrossRef]
- Huang, J.; Wang, A.; Huang, C.; Sun, Y.; Song, B.; Zhou, R.; Li, L. Generation of marker-free pbd-2 knock-in pigs using the crispr/cas9 and cre/loxp systems. Genes 2020, 11, 951. [Google Scholar] [CrossRef]
- Carlson, D.F.; Lancto, C.A.; Zang, B.; Kim, E.S.; Walton, M.; Oldeschulte, D.; Seabury, C.; Sonstegard, T.S.; Fahrenkrug, S.C. Production of hornless dairy cattle from genome-edited cell lines. Nat. Biotechnol. 2016, 34, 479–481. [Google Scholar] [CrossRef]
- Jeong, Y.H.; Kim, Y.J.; Kim, E.Y.; Kim, S.E.; Kim, J.; Park, M.J.; Lee, H.G.; Park, S.P.; Kang, M.J. Knock-in fibroblasts and transgenic blastocysts for expression of human fgf2 in the bovine β-casein gene locus using crispr/cas9 nuclease-mediated homologous recombination. Zygote 2016, 24, 442–456. [Google Scholar] [CrossRef]
- Smesny, S.; Milleit, B.; Schaefer, M.R.; Hesse, J.; Schlögelhofer, M.; Langbein, K.; Hipler, U.C.; Berger, M.; Cotter, D.R.; Sauer, H.; et al. Effects of omega-3 pufa on immune markers in adolescent individuals at ultra-high risk for psychosis—Results of the randomized controlled vienna omega-3 study. Schizophr. Res. 2017, 188, 110–117. [Google Scholar] [CrossRef]
- Matsuda, Y.; Habu, D.; Lee, S.; Kishida, S.; Osugi, H. Enteral diet enriched with ω-3 fatty acid improves oxygenation after thoracic esophagectomy for cancer: A randomized controlled trial. World J. Surg. 2017, 41, 1584–1594. [Google Scholar] [CrossRef] [PubMed]
- Bonafini, S.; Fava, C. Omega-3 fatty acids and cytochrome p450-derived eicosanoids in cardiovascular diseases: Which actions and interactions modulate hemodynamics? Prostaglandins Other Lipid Mediat. 2017, 128–129, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Ip, W.T.K.; Chandramouli, C.; Smith, J.A.; McLennan, P.L.; Pepe, S.; Delbridge, L.M.D. A small cohort omega-3 pufa supplement study: Implications of stratifying according to lipid membrane incorporation in cardiac surgical patients. Heart Lung Circ. 2017, 26, 846–855. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Mei, M.; Zhang, P.; Ma, K.; Song, G.; Ma, X.; Zhao, T.; Tang, B.; Ouyang, H.; Li, G.; et al. The generation of transgenic mice with fat1 and fad2 genes that have their own polyunsaturated fatty acid biosynthetic pathway. Cell. Physiol. Biochem. 2013, 32, 523–532. [Google Scholar] [CrossRef]
- Serini, S.; Calviello, G. Modulation of ras/erk and phosphoinositide signaling by long-chain n-3 pufa in breast cancer and their potential complementary role in combination with targeted drugs. Nutrients 2017, 9, 185. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Tang, Y.; Wang, S.; Zhou, J.; Zhou, J.; Lu, X.; Bai, X.; Wang, X.Y.; Chen, Z.; Zuo, D. Endogenous n-3 polyunsaturated fatty acids attenuate t cell-mediated hepatitis via autophagy activation. Front. Immunol. 2016, 7, 350. [Google Scholar] [CrossRef] [Green Version]
- van de Wetering, J.K.; Elfring, R.H.; Oosterlaken-Dijksterhuis, M.A.; Mol, J.A.; Haagsman, H.P.; Batenburg, J.J. Perinatal expression of igfbps in rat lung and its hormonal regulation in fetal lung explants. Am. J. Physiol. 1997, 273, L1174–L1181. [Google Scholar] [CrossRef]
- Froesch, E.R.; Schmid, C.; Schwander, J.; Zapf, J. Actions of insulin-like growth factors. Annu. Rev. Physiol. 1985, 47, 443–467. [Google Scholar] [CrossRef]
- Hammer, R.E.; Pursel, V.G.; Rexroad, C.E., Jr.; Wall, R.J.; Bolt, D.J.; Ebert, K.M.; Palmiter, R.D.; Brinster, R.L. Production of transgenic rabbits, sheep and pigs by microinjection. Nature 1985, 315, 680–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.L.; LeRoith, D. Insulin-like growth factor i is essential for postnatal growth in response to growth hormone. Endocrinology 1999, 140, 5178–5184. [Google Scholar] [CrossRef]
- Bodine, S.C.; Stitt, T.N.; Gonzalez, M.; Kline, W.O.; Stover, G.L.; Bauerlein, R.; Zlotchenko, E.; Scrimgeour, A.; Lawrence, J.C.; Glass, D.J.; et al. Akt/mtor pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001, 3, 1014–1019. [Google Scholar] [CrossRef]
- Grounds, M.D. Reasons for the degeneration of ageing skeletal muscle: A central role for igf-1 signalling. Biogerontology 2002, 3, 19–24. [Google Scholar] [CrossRef]
- Mourkioti, F.; Rosenthal, N. Igf-1, inflammation and stem cells: Interactions during muscle regeneration. Trends Immunol. 2005, 26, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Musarò, A.; McCullagh, K.; Paul, A.; Houghton, L.; Dobrowolny, G.; Molinaro, M.; Barton, E.R.; Sweeney, H.L.; Rosenthal, N. Localized igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat. Genet. 2001, 27, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Rommel, C.; Bodine, S.C.; Clarke, B.A.; Rossman, R.; Nunez, L.; Stitt, T.N.; Yancopoulos, G.D.; Glass, D.J. Mediation of igf-1-induced skeletal myotube hypertrophy by pi(3)k/akt/mtor and pi(3)k/akt/gsk3 pathways. Nat. Cell Biol. 2001, 3, 1009–1013. [Google Scholar] [CrossRef] [PubMed]
- Delafontaine, P.; Lou, H.; Harrison, D.G.; Bernstein, K.E. Sequence of a cdna encoding dog insulin-like growth factor i. Gene 1993, 130, 305–306. [Google Scholar] [CrossRef]
- Szymczak, A.L.; Vignali, D.A. Development of 2a peptide-based strategies in the design of multicistronic vectors. Expert Opin. Biol. Ther. 2005, 5, 627–638. [Google Scholar] [CrossRef]
- Xiong, Q.; Chen, J.; Li, F.L.; Zhao, S.; Wan, X.; Yang, X.; Li, J.; Luo, D.; Wang, Z.; Lv, X.; et al. Co-expression of mfat-1 and pig igf-1 genes by recombinant plasmids in modified chitosan nanoparticles and its synergistic effect on mouse immunity. Sci. Rep. 2017, 7, 17136. [Google Scholar] [CrossRef] [Green Version]
- Smith, H.K.; Matthews, K.G.; Oldham, J.M.; Jeanplong, F.; Falconer, S.J.; Bass, J.J.; Senna-Salerno, M.; Bracegirdle, J.W.; McMahon, C.D. Translational signalling, atrogenic and myogenic gene expression during unloading and reloading of skeletal muscle in myostatin-deficient mice. PLoS ONE 2014, 9, e94356. [Google Scholar] [CrossRef]
- Lai, L.; Kolber-Simonds, D.; Park, K.-W.; Cheong, H.-T.; Greenstein, J.L.; Im, G.-S.; Samuel, M.; Bonk, A.; Rieke, A.; Day, B.N.; et al. Production of α-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 2002, 295, 1089–1092. [Google Scholar] [CrossRef]
- Han, Y.M.; Park, J.M.; Cha, J.Y.; Jeong, M.; Go, E.J.; Hahm, K.B. Endogenous conversion of ω-6 to ω-3 polyunsaturated fatty acids in fat-1 mice attenuated intestinal polyposis by either inhibiting cox-2/β-catenin signaling or activating 15-pgdh/il-18. Int. J. Cancer 2016, 138, 2247–2256. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Nie, D.; Witt, W.T.; Chen, Q.; Shen, M.; Xie, H.; Lai, L.; Dai, Y.; Zhang, J. Expression of the fat-1 gene diminishes prostate cancer growth in vivo through enhancing apoptosis and inhibiting gsk-3 β phosphorylation. Mol. Cancer Ther. 2008, 7, 3203–3211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.M.; Kim, K.J.; Jeong, M.; Park, J.M.; Go, E.J.; Kang, J.X.; Hong, S.P.; Hahm, K.B. Suppressed helicobacter pylori-associated gastric tumorigenesis in fat-1 transgenic mice producing endogenous ω-3 polyunsaturated fatty acids. Oncotarget 2016, 7, 66606–66622. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Ouyang, H.; Yuan, H.; Li, J.; Xie, Z.; Wang, K.; Yu, T.; Liu, M.; Chen, X.; Tang, X.; et al. Site-specific fat-1 knock-in enables significant decrease of n-6pufas/n-3pufas ratio in pigs. G3 Genes Genomes Genet. 2018, 8, 1747–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Cui, M.L.; Nie, Y.W.; Dai, B.; Li, F.R.; Liu, D.J.; Liang, H.; Cang, M. Crispr/cas9-mediated specific integration of fat-1 at the goat mstn locus. FEBS J. 2018, 285, 2828–2839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, Y.; Xu, W.; Kan, Y.; Zhao, H.Y.; Zhou, Y.; Song, X.; Wu, J.; Xiong, J.; Goswami, D.; Yang, M.; et al. Extensive germline genome engineering in pigs. Nat. Biomed. Eng. 2021, 5, 134–143. [Google Scholar] [CrossRef]
- de Felipe, P.; Ryan, M.D. Targeting of proteins derived from self-processing polyproteins containing multiple signal sequences. Traffic 2004, 5, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Xu, Y.; Zhang, X.; Ding, C.; Huang, R.; Zhang, Z.; Lv, J.; Xie, X.; Chen, Y.; Li, Y.; et al. Crispr/cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 2015, 6, 363–372. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Sun, X.; Ding, F.; Zhang, K.; Zhao, R.; Li, S.; Li, R.; Tang, B.; Zhang, L.; Liu, Y.; et al. Removal of selectable marker gene from fibroblast cells in transgenic cloned cattle by transient expression of cre recombinase and subsequent effects on recloned embryo development. Theriogenology 2009, 72, 535–541. [Google Scholar] [CrossRef]
- Kang, J.X.; Wang, J.; Wu, L.; Kang, Z.B. Transgenic mice: Fat-1 mice convert n-6 to n-3 fatty acids. Nature 2004, 427, 504. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.; Kang, J.X.; Li, R.; Wang, J.; Witt, W.T.; Yong, H.Y.; Hao, Y.; Wax, D.M.; Murphy, C.N.; Rieke, A.; et al. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat. Biotechnol. 2006, 24, 435–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandy, S.; Srivastava, V. Site-specific gene integration in rice genome mediated by the flp-frt recombination system. Plant. Biotechnol. J. 2011, 9, 713–721. [Google Scholar] [CrossRef]
- Obayashi, H.; Kawabe, Y.; Makitsubo, H.; Watanabe, R.; Kameyama, Y.; Huang, S.; Takenouchi, Y.; Ito, A.; Kamihira, M. Accumulative gene integration into a pre-determined site using cre/loxp. J. Biosci. Bioeng. 2012, 113, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Wang, W.; Wu, H.; Chen, C.; Shen, D.; Wang, S.; Chen, W.; Zhang, L.; Chan, S.; Song, C. Changes in skeletal muscle and body weight on sleeping beauty transposon-mediated transgenic mice overexpressing pig migf-1. Biochem. Genet. 2018, 56, 341–355. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Dong, Y.; Meng, Q.; Zhao, Y.; Li, N. Incorporation of a skeletal muscle-specific enhancer in the regulatory region of igf1 upregulates igf1 expression and induces skeletal muscle hypertrophy. Sci. Rep. 2018, 8, 2781. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, Y.; Dou, H.; Yin, J.; Chen, Y.; Pang, X.; Vajta, G.; Bolund, L.; Du, Y.; Ma, R.Z. Handmade cloned transgenic piglets expressing the nematode fat-1 gene. Cell Reprogram. 2012, 14, 258–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Fatty Acids | Positive Pigs | Wild-Type Pigs |
|---|---|---|
| n-3 PUFAs | ||
| C22:5(DPA) | 22.195 ± 0.064221 *** | 12.496 ± 0.053226 *** |
| C22:6(DHA) | 12.042 ± 0.041313 *** | 5.750 ± 0.038649 *** |
| C20:5(EPA) | 39.160 ± 0.48597 *** | 23.523 ± 0.002471 *** |
| C18:3(ALA) | 18.553 ± 0.09573 *** | 7.612 ± 0.021941 *** |
| Total n-3 | 91.951 ± 0.079628 *** | 49.381 ± 0.029869 *** |
| n-6 PUFAs | ||
| C18:2(LA) | 138.318 ± 0.034776 *** | 243.525 ± 0.066764 *** |
| C20:4(AA) | 148.747 ± 0.058109 *** | 101.243 ± 0.094351 *** |
| Total n-6 | 287.065 ± 0.010417 *** | 344.768 ± 0.080017 *** |
| n-6/n-3 ratio | 3.122 | 6.982 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, W.; Li, M.; Qi, Y.; Wang, Y.; Chen, Y.; Liu, Y.; Li, L.; Ouyang, H.; Pang, D. CRISPR/Cas9-Mediated Specific Integration of Fat-1 and IGF-1 at the pRosa26 Locus. Genes 2021, 12, 1027. https://doi.org/10.3390/genes12071027
You W, Li M, Qi Y, Wang Y, Chen Y, Liu Y, Li L, Ouyang H, Pang D. CRISPR/Cas9-Mediated Specific Integration of Fat-1 and IGF-1 at the pRosa26 Locus. Genes. 2021; 12(7):1027. https://doi.org/10.3390/genes12071027
Chicago/Turabian StyleYou, Wenni, Mengjing Li, Yilin Qi, Yanbing Wang, Yiwu Chen, Ying Liu, Li Li, Hongsheng Ouyang, and Daxin Pang. 2021. "CRISPR/Cas9-Mediated Specific Integration of Fat-1 and IGF-1 at the pRosa26 Locus" Genes 12, no. 7: 1027. https://doi.org/10.3390/genes12071027
APA StyleYou, W., Li, M., Qi, Y., Wang, Y., Chen, Y., Liu, Y., Li, L., Ouyang, H., & Pang, D. (2021). CRISPR/Cas9-Mediated Specific Integration of Fat-1 and IGF-1 at the pRosa26 Locus. Genes, 12(7), 1027. https://doi.org/10.3390/genes12071027