Nature and Nurture: Genotype-Dependent Differential Responses of Root Architecture to Agar and Soil Environments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Constructs
2.2. Plant Growth Conditions and Experimental Set-Up
2.3. Data Acquisition
2.4. Data and Statistical Analysis
3. Results
3.1. plt CRISPR Mutants Resemble Described T-DNA Lines
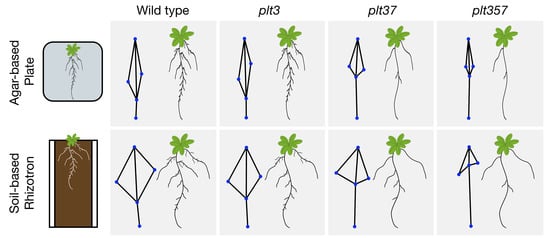
3.2. RSA Shape of plt3plt7 and plt3plt5plt7 Was Distinct from Other Genotypes in Plates and Rhizotrons
3.3. RSA Traits Differed between Plates and Rhizotrons, and Responses Were Genotype Dependent
3.4. Differential RSA Responses to Plates and Rhizotrons Not Limited to PLT Gene Family
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lynch, J. Root Architecture and Plant. Productivity. Plant Physiol. 1995, 109, 7–13. [Google Scholar] [CrossRef]
- Rogers, E.D.; Benfey, P.N. Regulation of plant root system architecture: Implications for crop advancement. Curr. Opin. Biotechnol. 2015, 32, 93–98. [Google Scholar] [CrossRef]
- Ge, Y.; Fang, X.; Liu, W.; Sheng, L.; Xu, L. Adventitious lateral rooting: The plasticity of root system architecture. Physiol. Plant. 2018, 165, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Rosas, U.; Cibrian-Jaramillo, A.; Ristova, D.; Banta, J.A.; Gifford, M.L.; Fan, A.H.; Zhou, R.W.; Kim, G.J.; Krouk, G.; Birnbaum, K.D.; et al. Integration of responses within and across Arabidopsis natural accessions uncovers loci controlling root sys-tems architecture. Proc. Natl. Acad. Sci. USA 2013, 110, 15133–15138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokawa, K.; Kagenishi, T.; Baluška, F. Root photomorphogenesis in laboratory-maintained Arabidopsis seedlings. Trends Plant Sci. 2013, 18, 117–119. [Google Scholar] [CrossRef]
- Nagel, K.A.; Putz, A.; Gilmer, F.; Heinz, K.; Fischbach, A.; Pfeifer, J.; Faget, M.; Blossfeld, S.; Ernst, M.; Dimaki, C.; et al. GROWSCREEN-Rhizo is a novel phenotyping robot enabling simultaneous measurements of root and shoot growth for plants grown in soil-filled rhizotrons. Funct. Plant Biol. 2012, 39, 891–904. [Google Scholar] [CrossRef] [Green Version]
- Devienne-Barret, F.; Richard-Molard, C.; Chelle, M.; Maury, O.; Ney, B. Ara-Rhizotron: An Effective Culture System to Study Simultaneously Root and Shoot Development of Arabidopsis. Plant Soil 2006, 280, 253–266. [Google Scholar] [CrossRef]
- Mooney, S.J.; Pridmore, T.P.; Helliwell, J.R.; Bennett, M.J. Developing X-ray Computed Tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil 2012, 352, 1–22. [Google Scholar] [CrossRef]
- Downie, H.; Holden, N.; Otten, W.; Spiers, A.; Valentine, T.A.; Dupuy, L.X. Transparent Soil for Imaging the Rhizosphere. PLoS ONE 2012, 7, e44276. [Google Scholar] [CrossRef]
- Morris, E.; Griffiths, M.; Golebiowska, A.; Mairhofer, S.; Burr-Hersey, J.; Goh, T.; von Wangenheim, D.; Atkinson, B.; Sturrock, C.J.; Lynch, J.P.; et al. Shaping 3D Root System Architecture. Curr. Biol. 2017, 27, R919–R930. [Google Scholar] [CrossRef] [Green Version]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.-S.; Amasino, R.; Scheres, B. The PLETHORA Genes Mediate Patterning of the Arabidopsis Root Stem Cell Niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galinha, C.; Hofhuis, H.; Luijten, M.; Willemsen, V.; Blilou, I.; Heidstra, R.; Scheres, B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nat. Cell Biol. 2007, 449, 1053–1057. [Google Scholar] [CrossRef] [Green Version]
- Hofhuis, H.; Laskowski, M.; Du, Y.; Prasad, K.; Grigg, S.; Pinon, V.; Scheres, B. Phyllotaxis and Rhizotaxis in Arabidopsis Are Modified by Three PLETHORA Transcription Factors. Curr. Biol. 2013, 23, 956–962. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Scheres, B. PLETHORA transcription factors orchestrate de novo organ patterning during Arabidopsis lateral root out-growth. Proc. Natl. Acad. Sci. USA 2017, 114, 11709–11714. [Google Scholar] [CrossRef] [Green Version]
- Engler, C.; Marillonnet, S. Golden Gate Cloning. Methods Mol. Biol. 2014, 1116, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.; Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef]
- Shimada, T.L.; Shimada, T.; Hara-Nishimura, I. A rapid and non-destructive screenable marker, FAST, for identifying trans-formed seeds of Arabidopsis thaliana. Plant J. 2010, 61, 519–528. [Google Scholar] [CrossRef]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering inArabidopsis thaliana. Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Fukaki, H.; Wysocka-Diller, J.; Kato, T.; Fujisawa, H.; Benfey, P.N.; Tasaka, M. Genetic evidence that the endodermis is essential for shoot gravitropism inArabidopsis thaliana. Plant J. 1998, 14, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Klingenberg, C.P. MorphoJ: An integrated software package for geometric morphometrics. Mol. Ecol. Resour. 2010, 11, 353–357. [Google Scholar] [CrossRef]
- Aceves-García, P.; Álvarez-Buylla, E.R.; Garay-Arroyo, A.; García-Ponce, B.; Muñoz, R.; Sánchez, M.D.L.P. Root Architecture Diversity and Meristem Dynamics in Different Populations of Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 858. [Google Scholar] [CrossRef] [Green Version]
- Rohlf, F.J. The tps series of software. Hystrix Ital. J. Mammal. 2015, 26, 9–12. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Lobet, G.; Pagès, L.; Draye, X. A Novel Image-Analysis Toolbox Enabling Quantitative Analysis of Root System Architecture. Plant Physiol. 2011, 157, 29–39. [Google Scholar] [CrossRef] [Green Version]
- Benfey, P.N.; Linstead, P.J.; Roberts, K.; Schiefelbein, J.W.; Hauser, M.T.; Aeschbacher, R.A. Root development in Ara-bidopsis: Four mutants with dramatically altered root morphogenesis. Development 1993, 119, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Di Laurenzio, L.; Wysocka-Diller, J.; Malamy, J.E.; Pysh, L.; Helariutta, Y.; Freshour, G.; Hahn, M.; Feldmann, K.A.; Benfey, P.N. The SCARECROW Gene Regulates an Asymmetric Cell Division That Is Essential for Generating the Radial Organization of the Arabidopsis Root. Cell 1996, 86, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Helariutta, Y.; Fukaki, H.; Wysocka-Diller, J.; Nakajima, K.; Jung, J.; Sena, G.; Hauser, M.-T.; Benfey, P.N. The SHORT-ROOT Gene Controls Radial Patterning of the Arabidopsis Root through Radial Signaling. Cell 2000, 101, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Scheres, B.; Di Laurenzio, L.; Willemsen, V.; Hauser, M.-T.; Janmaat, K.; Weisbeek, P.; Benfey, P.N. Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 1995, 121, 53–62. [Google Scholar] [CrossRef]
- Sabatini, S.; Heidstra, R.; Wildwater, M.; Scheres, B. SCARECROW is involved in positioning the stem cell niche in the Ara-bidopsis root meristem. Genes Dev. 2003, 17, 354–358. [Google Scholar] [CrossRef] [Green Version]
- Goh, T.; Toyokura, K.; Wells, D.M.; Swarup, K.; Yamamoto, M.; Mimura, T.; Weijers, D.; Fukaki, H.; Laplaze, L.; Bennett, M.J.; et al. Quiescent center initiation in the Arabidopsis lateral root primordia is dependent on the SCARECROW transcription factor. Development 2016, 143, 3363–3371. [Google Scholar] [CrossRef] [Green Version]
- Lucas, M.; Swarup, R.; Paponov, I.A.; Swarup, K.; Casimiro, I.; Lake, D.; Peret, B.; Zappala, S.; Mairhofer, S.; Whitworth, M.; et al. SHORT-ROOT Regulates Primary, Lateral, and Adventitious Root Development in Arabidopsis. Plant Physiol. 2011, 155, 384–398. [Google Scholar] [CrossRef] [Green Version]
- Rellán-Álvarez, R.; Lobet, G.; Lindner, H.; Pradier, P.-L.; Sebastian, J.L.; Yee, M.-C.; Geng, Y.; Trontin, C.; LaRue, T.; Schrager-Lavelle, A.; et al. GLO-Roots: An imaging platform enabling multidimensional characterization of soil-grown root systems. eLife 2015, 4, e07597. [Google Scholar] [CrossRef]
- Gandullo, J.; Ahmad, S.; Darwish, E.; Karlova, R.; Testerink, C. Phenotyping Tomato Root Developmental Plasticity in Re-sponse to Salinity in Soil Rhizotrons. Plant Phenomics 2021, 2021, 2760532. [Google Scholar] [CrossRef]
- Jiang, N.; Floro, E.; Bray, A.L.; Laws, B.; Duncan, K.E.; Topp, C.N. Three-Dimensional Time-Lapse Analysis Reveals Mul-tiscale Relationships in Maize Root Systems with Contrasting Architectures. Plant Cell 2019, 31, 1708–1722. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kerstens, M.; Hesen, V.; Yalamanchili, K.; Bimbo, A.; Grigg, S.; Opdenacker, D.; Beeckman, T.; Heidstra, R.; Willemsen, V. Nature and Nurture: Genotype-Dependent Differential Responses of Root Architecture to Agar and Soil Environments. Genes 2021, 12, 1028. https://doi.org/10.3390/genes12071028
Kerstens M, Hesen V, Yalamanchili K, Bimbo A, Grigg S, Opdenacker D, Beeckman T, Heidstra R, Willemsen V. Nature and Nurture: Genotype-Dependent Differential Responses of Root Architecture to Agar and Soil Environments. Genes. 2021; 12(7):1028. https://doi.org/10.3390/genes12071028
Chicago/Turabian StyleKerstens, Merijn, Vera Hesen, Kavya Yalamanchili, Andrea Bimbo, Stephen Grigg, Davy Opdenacker, Tom Beeckman, Renze Heidstra, and Viola Willemsen. 2021. "Nature and Nurture: Genotype-Dependent Differential Responses of Root Architecture to Agar and Soil Environments" Genes 12, no. 7: 1028. https://doi.org/10.3390/genes12071028
APA StyleKerstens, M., Hesen, V., Yalamanchili, K., Bimbo, A., Grigg, S., Opdenacker, D., Beeckman, T., Heidstra, R., & Willemsen, V. (2021). Nature and Nurture: Genotype-Dependent Differential Responses of Root Architecture to Agar and Soil Environments. Genes, 12(7), 1028. https://doi.org/10.3390/genes12071028