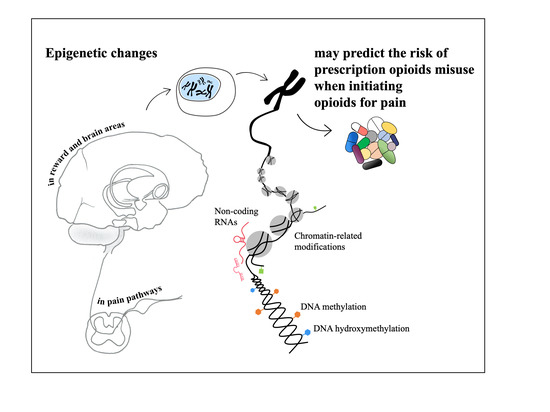
Epigenetic Alterations in Prescription Opioid Misuse: New Strategies for Precision Pain Management
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Prescription Opioids Pain Relievers
3.2. Epigenetics and Prescription Opioids
3.2.1. Animal Models’ Studies
3.2.2. Human Studies
3.3. Intergenerational and Transgenerational Epigenetic Effect of Prescription Opioids
3.4. What Can We Learn from Heroin Epigenetics?
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dowell, D.; Haegerich, T.M.; Chou, R. CDC Guideline for Prescribing Opioids for Chronic Pain—United States, 2016. Recomm. Rep. 2016, 65, 1–49. [Google Scholar] [CrossRef]
- Vowles, K.E.; McEntee, M.L.; Julnes, P.S.; Frohe, T.; Ney, J.P.; van der Goes, D.N. Rates of opioid misuse, abuse, and addiction in chronic pain: A systematic review and data synthesis. Pain 2015, 156, 569–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- UNODC. World Drug Report 2020 (United Nations Publication, Sales No. E.20.XI.6); UNODC: Vienna, Austria, 2020. [Google Scholar]
- Fishbain, D.A.; Cole, B.; Lewis, J.; Rosomoff, H.L.; Rosomoff, R.S. What percentage of chronic nonmalignant pain patients exposed to chronic opioid analgesic therapy develop abuse/addiction and/or aberrant drug-related behaviors? A structured evidence-based review. Pain Med. 2008, 9, 444–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centers for Disease Control and Prevention (CDC) Wide-Ranging Online Data for Epidemiologic Research (WONDER). National Center for Health Statistics. Available online: http://wonder.cdc.gov (accessed on 30 September 2020).
- Trescot, A.M.; Datta, S.; Lee, M.; Hansen, H. Opioid pharmacology. Pain Physician 2008, 11, S133–S153. [Google Scholar] [CrossRef]
- Manhapra, A.; Becker, W.C. Pain and Addiction: An Integrative Therapeutic Approach. Med. Clin. N. Am. 2018, 102, 745–763. [Google Scholar] [CrossRef]
- Biernikiewicz, M.; Taieb, V.; Toumi, M. Characteristics of doctor-shoppers: A systematic literature review. J. Mark. Access Health Policy 2019, 7, 1595953. [Google Scholar] [CrossRef] [Green Version]
- Volkow, N.; Benveniste, H.; McLellan, A.T. Use and Misuse of Opioids in Chronic Pain. Annu. Rev. Med. 2018, 69, 451–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voon, P.; Karamouzian, M.; Kerr, T. Chronic pain and opioid misuse: A review of reviews. Subst. Abuse Treat. Prev. Policy 2017, 12, 36. [Google Scholar] [CrossRef] [Green Version]
- Vieira, C.M.P.; Fragoso, R.M.; Pereira, D.; Medeiros, R. Pain polymorphisms and opioids: An evidence based review. Mol. Med. Rep. 2019, 19, 1423–1434. [Google Scholar] [CrossRef] [Green Version]
- Klimas, J.; Gorfinkel, L.; Fairbairn, N.; Amato, L.; Ahamad, K.; Nolan, S.; Simel, D.L.; Wood, E. Strategies to Identify Patient Risks of Prescription Opioid Addiction When Initiating Opioids for Pain: A Systematic Review. JAMA Netw. Open 2019, 2, e193365. [Google Scholar] [CrossRef]
- Merrick, M.T.; Ford, D.C.; Haegerich, T.M.; Simon, T. Adverse Childhood Experiences Increase Risk for Prescription Opioid Misuse. J. Prim. Prev. 2020, 41, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Miyake, K.; Hirasawa, T. Epigenetic understanding of gene-environment interactions in psychiatric disorders: A new concept of clinical genetics. Clin. Epigenet. 2012, 4, 1. [Google Scholar] [CrossRef] [Green Version]
- Browne, C.J.; Godino, A.; Salery, M.; Nestler, E.J. Epigenetic Mechanisms of Opioid Addiction. Biol. Psychiatry 2020, 87, 22–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilardi, F.; Augsburger, M.; Thomas, A. Will Widespread Synthetic Opioid Consumption Induce Epigenetic Consequences in Future Generations? Front. Pharmacol. 2018, 9, 702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapocik, J.D.; Letwin, N.; Mayo, C.L.; Frank, B.; Luu, T.; Achinike, O.; House, C.; Williams, R.; Elmer, G.I.; Lee, N.H. Identification of candidate genes and gene networks specifically associated with analgesic tolerance to morphine. J. Neurosci. 2009, 29, 5295–5307. [Google Scholar] [CrossRef]
- Sun, H.; Maze, I.; Dietz, D.M.; Scobie, K.N.; Kennedy, P.J.; Damez-Werno, D.; Neve, R.L.; Zachariou, V.; Shen, L.; Nestler, E.J. Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. J. Neurosci. 2012, 32, 17454–17464. [Google Scholar] [CrossRef]
- Zhang, Z.; Tao, W.; Hou, Y.-Y.; Wang, W.; Kenny, P.J.; Pan, Z.Z. MeCP2 repression of G9a in regulation of pain and morphine reward. J. Neurosci. 2014, 34, 9076–9087. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.-Y.; Cai, Y.-Q.; Pan, Z.Z. Persistent pain maintains morphine-seeking behavior after morphine withdrawal through reduced MeCP2 repression of GluA1 in rat central amygdala. J. Neurosci. 2015, 35, 3689–3700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Chen, S.-R.; Laumet, G.; Chen, H.; Pan, H.-L. Nerve Injury Diminishes Opioid Analgesia through Lysine Methyltransferase-mediated Transcriptional Repression of μ-Opioid Receptors in Primary Sensory Neurons. J. Biol. Chem. 2016, 291, 8475–8485. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Mao, Y.; Liang, L.; Wu, S.; Yuan, J.; Mo, K.; Cai, W.; Mao, Q.; Cao, J.; Bekker, A.; et al. The transcription factor C/EBPβ in the dorsal root ganglion contributes to peripheral nerve trauma-induced nociceptive hypersensitivity. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef]
- Wang, Y.; Lai, J.; Cui, H.; Zhu, Y.; Zhao, B.; Wang, W.; Wei, S. Inhibition of histone deacetylase in the basolateral amygdala facilitates morphine context-associated memory formation in rats. J. Mol. Neurosci. 2015, 55, 269–278. [Google Scholar] [CrossRef]
- Barrow, T.M.; Byun, H.-M.; Li, X.; Smart, C.; Wang, Y.-X.; Zhang, Y.; Baccarelli, A.A.; Guo, L. The effect of morphine upon DNA methylation in ten regions of the rat brain. Epigenetics 2017, 12, 1038–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Q.; Hwang, C.K.; Zheng, H.; Wagley, Y.; Lin, H.-Y.; Kim, D.K.; Law, P.-Y.; Loh, H.H.; Wei, L.-N. MicroRNA 339 down-regulates μ-opioid receptor at the post-transcriptional level in response to opioid treatment. FASEB J. 2013, 27, 522–535. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.-M.; Cao, S.-B.; Zhang, H.-L.; Lyu, D.-M.; Chen, L.-P.; Xu, H.; Pan, Z.-Q.; Shen, W. Downregulation of miR-219 enhances brain-derived neurotrophic factor production in mouse dorsal root ganglia to mediate morphine analgesic tolerance by upregulating CaMKIIγ. Mol. Pain 2016, 12, 1744806916666283. [Google Scholar] [CrossRef]
- Li, H.; Tao, R.; Wang, J.; Xia, L. Upregulation of miR-375 level ameliorates morphine analgesic tolerance in mouse dorsal root ganglia by inhibiting the JAK2/STAT3 pathway. J. Pain Res. 2017, 10, 1279–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpenter, M.D.; Manners, M.T.; Heller, E.A.; Blendy, J.A. Adolescent oxycodone exposure inhibits withdrawal-induced expression of genes associated with the dopamine transmission. Addict. Biol. 2020, e12994. [Google Scholar] [CrossRef]
- Yuferov, V.; Zhang, Y.; Liang, Y.; Zhao, C.; Randesi, M.; Kreek, M.J. Oxycodone Self-Administration Induces Alterations in Expression of Integrin, Semaphorin and Ephrin Genes in the Mouse Striatum. Front. Psychiatry 2018, 9, 257. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liang, Y.; Levran, O.; Randesi, M.; Yuferov, V.; Zhao, C.; Kreek, M.J. Alterations of expression of inflammation/immune-related genes in the dorsal and ventral striatum of adult C57BL/6J mice following chronic oxycodone self-administration: A RNA sequencing study. Psychopharmacology 2017, 234, 2259–2275. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.-Y.; Shi, G.; Zhao, P. Reversal of oxycodone conditioned place preference by oxytocin: Promoting global DNA methylation in the hippocampus. Neuropharmacology 2019, 160, 107778. [Google Scholar] [CrossRef]
- Fan, X.-Y.; Shi, G.; Zhao, P. Methylation in Syn and Psd95 genes underlie the inhibitory effect of oxytocin on oxycodone-induced conditioned place preference. Eur. Neuropsychopharmacol. 2019, 29, 1464–1475. [Google Scholar] [CrossRef] [PubMed]
- Montalvo-Ortiz, J.L.; Cheng, Z.; Kranzler, H.R.; Zhang, H.; Gelernter, J. Author Correction: Genomewide Study of Epigenetic Biomarkers of Opioid Dependence in European- American Women. Sci. Rep. 2019, 9, 18774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandoval-Sierra, J.V.; Salgado García, F.I.; Brooks, J.H.; Derefinko, K.J.; Mozhui, K. Effect of short-term prescription opioids on DNA methylation of the OPRM1 promoter. Clin. Epigenet. 2020, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Kringel, D.; Kaunisto, M.A.; Kalso, E.; Lötsch, J. Machine-learned analysis of global and glial/opioid intersection-related DNA methylation in patients with persistent pain after breast cancer surgery. Clin. Epigenet. 2019, 11, 167. [Google Scholar] [CrossRef] [PubMed]
- Devarapalli, M.; Leonard, M.; Briyal, S.; Stefanov, G.; Puppala, B.L.; Schweig, L.; Gulati, A. Prenatal Oxycodone Exposure Alters CNS Endothelin Receptor Expression in Neonatal Rats. Drug Res. (Stuttg) 2016, 66, 246–250. [Google Scholar] [CrossRef]
- Wachman, E.M.; Hayes, M.J.; Lester, B.M.; Terrin, N.; Brown, M.S.; Nielsen, D.A.; Davis, J.M. Epigenetic variation in the mu-opioid receptor gene in infants with neonatal abstinence syndrome. J. Pediatr. 2014, 165, 472–478. [Google Scholar] [CrossRef] [Green Version]
- Shahjin, F.; Guda, R.S.; Schaal, V.L.; Odegaard, K.; Clark, A.; Gowen, A.; Xiao, P.; Lisco, S.J.; Pendyala, G.; Yelamanchili, S.V. Brain-Derived Extracellular Vesicle microRNA Signatures Associated with In Utero and Postnatal Oxycodone Exposure. Cells 2019, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Byrnes, J.J.; Johnson, N.L.; Carini, L.M.; Byrnes, E.M. Multigenerational effects of adolescent morphine exposure on dopamine D2 receptor function. Psychopharmacology 2013, 227, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Vassoler, F.M.; Oliver, D.J.; Wyse, C.; Blau, A.; Shtutman, M.; Turner, J.R.; Byrnes, E.M. Transgenerational attenuation of opioid self-administration as a consequence of adolescent morphine exposure. Neuropharmacology 2017, 113, 271–280. [Google Scholar] [CrossRef] [Green Version]
- Rosenblum, A.; Marsch, L.A.; Joseph, H.; Portenoy, R.K. Opioids and the treatment of chronic pain: Controversies, current status, and future directions. Exp. Clin. Psychopharmacol. 2008, 16, 405–416. [Google Scholar] [CrossRef]
- O’Brien, T.; Christrup, L.L.; Drewes, A.M.; Fallon, M.T.; Kress, H.G.; McQuay, H.J.; Mikus, G.; Morlion, B.J.; Perez-Cajaraville, J.; Pogatzki-Zahn, E.; et al. European Pain Federation position paper on appropriate opioid use in chronic pain management. Eur. J. Pain 2017, 21, 3–19. [Google Scholar] [CrossRef]
- Elman, I.; Borsook, D. Common Brain Mechanisms of Chronic Pain and Addiction. Neuron 2016, 89, 11–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballantyne, J.C. Opioids for the Treatment of Chronic Pain: Mistakes Made, Lessons Learned, and Future Directions. Anesth. Analg. 2017, 125, 1769–1778. [Google Scholar] [CrossRef] [PubMed]
- Deans, C.; Maggert, K.A. What do you mean, “epigenetic”? Genetics 2015, 199, 887–896. [Google Scholar] [CrossRef] [Green Version]
- Kundu, T.K.; Rao, M.R. CpG islands in chromatin organization and gene expression. J. Biochem. 1999, 125, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.R.; Yarychkivska, O.; Boulard, M.; Bestor, T.H. DNA methylation and DNA methyltransferases. Epigenet. Chromatin 2017, 10, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doehring, A.; Oertel, B.G.; Sittl, R.; Lötsch, J. Chronic opioid use is associated with increased DNA methylation correlating with increased clinical pain. Pain 2013, 154, 15–23. [Google Scholar] [CrossRef]
- Zheng, H.; Xie, W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 2019, 20, 535–550. [Google Scholar] [CrossRef]
- McDaid, J.; Dallimore, J.E.; Mackie, A.R.; Napier, T.C. Changes in accumbal and pallidal pCREB and deltaFosB in morphine-sensitized rats: Correlations with receptor-evoked electrophysiological measures in the ventral pallidum. Neuropsychopharmacology 2006, 31, 1212–1226. [Google Scholar] [CrossRef] [Green Version]
- Hwang, C.K.; Kim, C.S.; Kim, D.K.; Law, P.-Y.; Wei, L.-N.; Loh, H.H. Up-regulation of the mu-opioid receptor gene is mediated through chromatin remodeling and transcriptional factors in differentiated neuronal cells. Mol. Pharmacol. 2010, 78, 58–68. [Google Scholar] [CrossRef] [Green Version]
- Martin, J.A.; Caccamise, A.; Werner, C.T.; Viswanathan, R.; Polanco, J.J.; Stewart, A.F.; Thomas, S.A.; Sim, F.J.; Dietz, D.M. A Novel Role for Oligodendrocyte Precursor Cells (OPCs) and Sox10 in Mediating Cellular and Behavioral Responses to Heroin. Neuropsychopharmacology 2018, 43, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Nestler, E.J. Epigenetic mechanisms of drug addiction. Neuropharmacology 2014, 76 Pt B, 259–268. [Google Scholar] [CrossRef] [Green Version]
- DiStefano, J.K. The Emerging Role of Long Noncoding RNAs in Human Disease. Methods Mol. Biol. 2018, 1706, 91–110. [Google Scholar] [CrossRef]
- Yan, B.; Hu, Z.; Yao, W.; Le, Q.; Xu, B.; Liu, X.; Ma, L. MiR-218 targets MeCP2 and inhibits heroin seeking behavior. Sci. Rep. 2017, 7, 40413. [Google Scholar] [CrossRef] [Green Version]
- Niederberger, E.; Resch, E.; Parnham, M.J.; Geisslinger, G. Drugging the pain epigenome. Nat. Rev. Neurol. 2017, 13, 434–447. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, G.; Edelman, E.J.; Barry, D.T.; Becker, W.C.; Cerdá, M.; Crystal, S.; Gaither, J.R.; Gordon, A.J.; Gordon, K.S.; Kerns, R.D.; et al. Non-medical use of prescription opioids is associated with heroin initiation among US veterans: A prospective cohort study. Addiction 2016, 111, 2021–2031. [Google Scholar] [CrossRef]
- Krashin, D.; Murinova, N.; Sullivan, M. Challenges to Treatment of Chronic Pain and Addiction During the “Opioid Crisis”. Curr. Pain Headache Rep. 2016, 20, 65. [Google Scholar] [CrossRef]
- Chou, R.; Turner, J.A.; Devine, E.B.; Hansen, R.N.; Sullivan, S.D.; Blazina, I.; Dana, T.; Bougatsos, C.; Deyo, R.A. The effectiveness and risks of long-term opioid therapy for chronic pain: A systematic review for a National Institutes of Health Pathways to Prevention Workshop. Ann. Intern. Med. 2015, 162, 276–286. [Google Scholar] [CrossRef] [Green Version]
- Neugebauer, V. Amygdala pain mechanisms. Pain Control 2015, 227, 261–284. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.K.; Hu, J.; Meng, Q.; Xia, Y.; Shapiro, P.S.; Reddy, S.P.M.; Platanias, L.C.; Lindner, D.J.; Johnson, P.F.; Pritchard, C.; et al. MEKK1 plays a critical role in activating the transcription factor C/EBP-β-dependent gene expression in response to IFN-γ. Proc. Natl. Acad. Sci. USA 2002, 99, 7945–7950. [Google Scholar] [CrossRef] [Green Version]
- Cata, J.P.; Gorur, A.; Yuan, X.; Berg, N.K.; Sood, A.K.; Eltzschig, H.K. Role of Micro-RNA for Pain After Surgery: Narrative Review of Animal and Human Studies. Anesth. Analg. 2020, 130, 1638–1652. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.N.; Volta, M.; Pidsley, R.; Lunnon, K.; Harris, R.A.; Dobson, R.; Schalkwyk, L.C.; Dixit, A.; Lovestone, S.; Troakes, C.; et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012, 13, R43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massart, R.; Dymov, S.; Millecamps, M.; Suderman, M.; Gregoire, S.; Koenigs, K.; Alvarado, S.; Tajerian, M.; Stone, L.S.; Szyf, M. Overlapping signatures of chronic pain in the DNA methylation landscape of prefrontal cortex and peripheral T cells. Sci. Rep. 2016, 6, 19615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, A.S.; Estécio, M.R.H.; Doshi, K.; Kondo, Y.; Tajara, E.H.; Issa, J.-P.J. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004, 32, e38. [Google Scholar] [CrossRef]
- Blake, G.E.; Watson, E.D. Unravelling the complex mechanisms of transgenerational epigenetic inheritance. Curr. Opin. Chem. Biol. 2016, 33, 101–107. [Google Scholar] [CrossRef] [Green Version]
- Heard, E.; Martienssen, R.A. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 2014, 157, 95–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmadian-Moghadam, H.; Sadat-Shirazi, M.-S.; Seifi, F.; Niknamfar, S.; Akbarabadi, A.; Toolee, H.; Zarrindast, M.-R. Transgenerational influence of parental morphine exposure on pain perception, anxiety-like behavior and passive avoidance memory among male and female offspring of Wistar rats. EXCLI J. 2019, 18, 1019–1036. [Google Scholar] [CrossRef] [PubMed]
- D’Agnelli, S.; Gerra, M.C.; Bignami, E.; Arendt-Nielsen, L. Exosomes as a new pain biomarker opportunity. Mol. Pain 2020, 16. [Google Scholar] [CrossRef]
- Albertson, D.N.; Schmidt, C.J.; Kapatos, G.; Bannon, M.J. Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropharmacology 2006, 31, 2304–2312. [Google Scholar] [CrossRef] [Green Version]
- Kozlenkov, A.; Jaffe, A.E.; Timashpolsky, A.; Apontes, P.; Rudchenko, S.; Barbu, M.; Byne, W.; Hurd, Y.L.; Horvath, S.; Dracheva, S. DNA Methylation Profiling of Human Prefrontal Cortex Neurons in Heroin Users Shows Significant Difference between Genomic Contexts of Hyper- and Hypomethylation and a Younger Epigenetic Age. Genes 2017, 8, 152. [Google Scholar] [CrossRef] [Green Version]
- Hong, Q.; Xu, W.; Lin, Z.; Liu, J.; Chen, W.; Zhu, H.; Lai, M.; Zhuang, D.; Xu, Z.; Fu, D.; et al. Role of GABRD Gene Methylation in the Nucleus Accumbens in Heroin-Seeking Behavior in Rats. Front. Pharmacol. 2020, 11, 612200. [Google Scholar] [CrossRef]
- Egervari, G.; Akpoyibo, D.; Rahman, T.; Fullard, J.F.; Callens, J.E.; Landry, J.A.; Ly, A.; Zhou, X.; Warren, N.; Hauberg, M.E.; et al. Chromatin accessibility mapping of the striatum identifies tyrosine kinase FYN as a therapeutic target for heroin use disorder. Nat. Commun. 2020, 11, 4634. [Google Scholar] [CrossRef]
- Levine, A.; Clemenza, K.; Weiss, S.; Bisaga, A.; Eitan, E.; Gerra, M.C.; Donnini, C.; Gerra, G.; Garcia, B. Discovering Non-Invasive Biomarkers Predictive of Opioid Use Disorder Treatment Response. CNS Spectr. 2021, 26, 173. [Google Scholar] [CrossRef]
- Sheng, J.; Lv, Z.G.; Wang, L.; Zhou, Y.; Hui, B. Histone H3 phosphoacetylation is critical for heroin-induced place preference. Neuroreport 2011, 22, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Hong, Q.; Liu, J.; Lin, Z.; Zhuang, D.; Xu, W.; Xu, Z.; Lai, M.; Zhu, H.; Zhou, W.; Liu, H. Histone 3 lysine 9 acetylation of BRG1 in the medial prefrontal cortex is associated with heroin self-administration in rats. Mol. Med. Rep. 2020, 21, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-S.; Xu, W.-J.; Zhu, H.-Q.; Gao, L.; Lai, M.-J.; Zhang, F.-Q.; Zhou, W.-H.; Liu, H.-F. Effects of histone deacetylase inhibitor sodium butyrate on heroin seeking behavior in the nucleus accumbens in rats. Brain Res. 2016, 1652, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Imperio, C.G.; McFalls, A.J.; Hadad, N.; Blanco-Berdugo, L.; Masser, D.R.; Colechio, E.M.; Coffey, A.A.; Bixler, G.V.; Stanford, D.R.; Vrana, K.E.; et al. Exposure to environmental enrichment attenuates addiction-like behavior and alters molecular effects of heroin self-administration in rats. Neuropharmacology 2018, 139, 26–40. [Google Scholar] [CrossRef]
- Fragou, D.; Chao, M.-R.; Hu, C.-W.; Nikolaou, K.; Kovatsi, L. Global DNA methylation levels in white blood cells of patients with chronic heroin use disorder. A prospective study. Toxicol. Rep. 2021, 8, 337–342. [Google Scholar] [CrossRef]
- Zhang, J.; Abdullah, J.M. The role of GluA1 in central nervous system disorders. Rev. Neurosci. 2013, 24, 499–505. [Google Scholar] [CrossRef]
- Anderson, E.M.; Sun, H.; Guzman, D.; Taniguchi, M.; Cowan, C.W.; Maze, I.; Nestler, E.J.; Self, D.W. Knockdown of the histone di-methyltransferase G9a in nucleus accumbens shell decreases cocaine self-administration, stress-induced reinstatement, and anxiety. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2019, 44, 1370–1376. [Google Scholar] [CrossRef]
- Castrén, E.; Antila, H. Neuronal plasticity and neurotrophic factors in drug responses. Mol. Psychiatry 2017, 22, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Webster, L.R. Risk Factors for Opioid-Use Disorder and Overdose. Anesth. Analg. 2017, 125, 1741–1748. [Google Scholar] [CrossRef]
- Wiss, D.A. A Biopsychosocial Overview of the Opioid Crisis: Considering Nutrition and Gastrointestinal Health. Front. Public Health 2019, 7, 193. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, M.C.; Case, L.K.; Ceko, M.; Cotton, V.A.; Gracely, J.L.; Low, L.A.; Pitcher, M.H.; Villemure, C. Effect of environment on the long-term consequences of chronic pain. Pain 2015, 156, S42–S49. [Google Scholar] [CrossRef] [PubMed]
- Contet, C.; Kieffer, B.L.; Befort, K. Mu opioid receptor: A gateway to drug addiction. Curr. Opin. Neurobiol. 2004, 14, 370–378. [Google Scholar] [CrossRef]
- Granstrem, O.; Adriani, W.; Shumilina, M.; Izykenova, G.; Dambinova, S.; Laviola, G. Specific changes in levels of autoantibodies to glutamate and opiate receptors induced by morphine administration in rats. Neurosci. Lett. 2006, 403, 1–5. [Google Scholar] [CrossRef]
- Capone, F.; Adriani, W.; Shumilina, M.; Izykenova, G.; Granstrem, O.; Dambinova, S.; Laviola, G. Autoantibodies against opioid or glutamate receptors are associated with changes in morphine reward and physical dependence in mice. Psychopharmacology 2008, 197, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Ognibene, E.; Adriani, W.; Granstrem, O.; Pieretti, S.; Laviola, G. Impulsivity-anxiety-related behavior and profiles of morphine-induced analgesia in heterozygous reeler mice. Brain Res. 2007, 1131, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Gapp, K.; Jawaid, A.; Sarkies, P.; Bohacek, J.; Pelczar, P.; Prados, J.; Farinelli, L.; Miska, E.; Mansuy, I.M. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 2014, 17, 667–669. [Google Scholar] [CrossRef] [Green Version]
- Radford, E.J.; Ito, M.; Shi, H.; Corish, J.A.; Yamazawa, K.; Isganaitis, E.; Seisenberger, S.; Hore, T.A.; Reik, W.; Erkek, S.; et al. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science 2014, 345, 1255903. [Google Scholar] [CrossRef] [Green Version]
- Padmanabhan, N.; Jia, D.; Geary-Joo, C.; Wu, X.; Ferguson-Smith, A.C.; Fung, E.; Bieda, M.C.; Snyder, F.F.; Gravel, R.A.; Cross, J.C.; et al. Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell 2013, 155, 81–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anway, M.D.; Leathers, C.; Skinner, M.K. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology 2006, 147, 5515–5523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egervari, G.; Landry, J.; Callens, J.; Fullard, J.F.; Roussos, P.; Keller, E.; Hurd, Y.L. Striatal H3K27 Acetylation Linked to Glutamatergic Gene Dysregulation in Human Heroin Abusers Holds Promise as Therapeutic Target. Biol. Psychiatry 2017, 81, 585–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Opioids | Tissues/Sample | Epigenetic Methods | Change | Animals | Findings | PMID | Authors |
|---|---|---|---|---|---|---|---|
| Morphine | Brain tissues (PAG, PFC, temporal lobe, and ventral striatum) | Microarray gene expression profiling and pattern matching | Gene expression | Adult male mice | The development of tolerance is influenced by a region in OPRM1 gene. The genes epigenetically modified by chronic morphine administration are mainly related to neuroadaptation. | 19386926 | Tapocik et al., 2009 [17] |
| Morphine | NAc | Chromatin immunoprecipitation followed by massive parallel sequencing | H3K9me2 distribution in NAc in the absence and presence of chronic morphine | 9 to 11-week-old C57BL/6J male mice or G9afl/fl mice | Chronic morphine decreases G9a expression and H3K9me2 at global level and in specific loci in mouse NAc. | 23197736 | Sun et al., 2012 [18] |
| Morphine | Central nucleus of amygdala | Chromatin immunoprecipitation | Gene and protein expression | Female mice with persistent and acute pain | Persistent pain and repeated morphine upregulate the transcriptional regulator MeCP2. MeCP2 enhances BDNF expression and represses G9a action and its repressive mark H3K9me2 in CeA. | 24990928 | Zhang et al., 2014 [19] |
| Morphine | Central nucleus of amygdala | Chromatin immunoprecipitation | Gene expression | Rat model of morphine self-administration | The repression of GluA1 function by MeCp2 is proposed as a mechanism for morphine-seeking behavior in pain experience. | 25716866 | Hou et al., 2015 [20] |
| Morphine | Dorsal root ganglia and spinal cord tissues | Quantitative RT-PCR, Western Immunoblotting and ChIP-PCR | Gene and protein expression, histone modifications analysis | Male Sprague-Dawley rats SNL (spinal nerve ligation) model | G9a contributes to transcriptional repression of MORs in primary sensory neurons in neuropathic pain. G9a inhibitors: potential treatment of chronic neuropathic pain | 26917724 | Zhang et al., 2016 [21] |
| Morphine | Dorsal root ganglia | Quantitative RT-PCR and Western Blot | Gene and protein expression | Adult male CD-1 mice | Neuropathic pain increases C/EBPβ expression. C/EPBβ activates the G9a gene, that epigenetically silences Kv1.2 and MOR genes. Blocking the induced increase in C/EBPβ in the DRG, morphine analgesia after CCI is improved. | 28698219 | Li et al., 2017 [22] |
| Morphine | Basolateral amygdala | Quantitative RT-PCR and Western Blot | Gene and protein expression | Male Sprague–Dawley | Increase in H3K14ac together with upregulation of the BDNF and FosB; and CREB activation. | 24829091 | Wang et al., 2015 [23] |
| Morphine | Rat brain regions | Pyrosequencing | DNA methylation (5mC) and global DNA 5-hydroxymethylation (5hmC) | Male Wistar rats | Acute and chronic exposure is associated with significantly decreased/increased 5mC at specific genes (BDNF, IL1B, IL6, NR3C1, COMT). Global 5hmC levels increase in the cerebral cortex, hippocampus, and hypothalamus, but decrease in the midbrain. | 29111854 | Barrow et al., 2017 [24] |
| Morphine, phentayl | Hippocampus | RNAseq | Gene and protein expression | Mice chronically treated with μ-opioid agonists | The increased expression of MiR-339-3p inhibits intracellular MOR biosynthesis and acts as a negative feedback modulator of MOR signals. | 23085997 | Wu et al., 2013 [25] |
| Morphine | Dorsal root ganglia | Quantitative RT-PCR and Western Blot | Gene and protein expression | Male CD-1 mice treated with morphine to establish systemic chronic tolerance to morphine anti-nociception | MiR-219 contributes to the development of chronic tolerance to morphine analgesia by targeting CaMKIIγ and enhancing CaMKIIγ-dependent brain-derived neurotrophic factor expression. | 27599867 | Hu et al., 2016 [26] |
| Morphine | Dorsal root ganglia | Quantitative RT-PCR and Western Blot | Gene and protein expression | Male CD-1 mice injected with morphine to elicit morphine tolerance | The increased BDNF expression is regulated by the miR-375 and JAK2/STAT3 pathway. Inhibition of this pathway decreases BDNF production, and thus, attenuated morphine tolerance. | 28603428 | Li et al., 2017 [27] |
| Oxycodone | Ventral tegmental area of the developing brain | Quantitative RT-PCR and chromatin immunoprecipitation | Gene expression and histone modifications analysis | Male offspring of C57Bl/6NTac mice | Adolescent oxycodone exposure increases the repressive mark H3K27me3, at key dopamine-related genes. | 33325096 | Carpenter et al., 2020 [28] |
| Oxycodone | Striatum (NAc and CPu) | RNAseq | Gene expression | Mice following extended 14-day oxycodone self-administration | Alterations in the expression of heterodimer receptor, integrins, semaphorins, semaphorin receptors, plexins, selective axon guidance genes. | 29946272 | Yuferov et al., 2018 [29] |
| Oxycodone | Dorsal striatum and ventral striatum | RNAseq | Gene expression | Adult male C57BL/6J mice underwent a 14-day oxycodone self-administration | Inflammation/immune genes have altered expression during chronic self-administration of oxycodone | 28653080 | Zhang et al., 2017 [30] |
| Oxycodone | Hippocampus | DNA ELISA Kit for total 5mC; quantitative RT-PCR | Global 5mC levels and gene expression | Male Sprague-Dawley rats | The global DNA hypomethylation induced by oxycodone can be reversed through oxytocin and could significantly attenuate the oxycodone rewarding effects. | 31526808 | Fan et al., 2019 [31] |
| Oxycodone | Ventral tegmental area | DNA ELISA Kit for total 5mC and OneStep qMethyl™ kit for gene-specific 5mC, quantitative RT-PCR, Western blotting | Global and specific 5mC levels and gene expression | Sprague-Dawley rats | Down-regulation of DNMT1 and up-regulation of TET1-3 lead to a decrease in global 5mC levels and differential demethylation at exon 1 of SYN and exon 2 of PSD95. | 31735530 | Fan et al., 2019 [32] |
| Opioids | Tissues | Epigenetic Methods | Change | Sample | Findings | PMID | Authors |
|---|---|---|---|---|---|---|---|
| Opioids | Whole blood | Bisulfite modification and Array-based genome-wide DNA methylation assay | DNA methylation at specific CpG sites | 140 opioid dependence cases and 80 opioid-exposed controls | Three genome-wide significant differentially methylated CpGs map to genes involved in chromatin remodeling, DNA binding, cell survival, and cell projection (PARG, RERE, and CFAP77 genes). | 31801960 | Montalvo-Ortiz et al., 2019 [33] |
| Opioid medication self-administration (hydrocodone, oxycodone, and codeine: 5–30 mg) | Saliva collected at 3 time points | Genome-wide DNA methylation assay and candidate approach | DNA methylation at OPRM1 gene promoter | 33 opioid-naïve participants who underwent standard dental surgery | Hypermethylation of the OPRM1 promoter is measured in response to opioid use, and such epigenetic restructuring can be induced even by short-term use of therapeutic opioids. | 32493461 | Sandoval-Sierra et al., 2020 [34] |
| Remifentanil, oxycodone, codeine | Whole blood | Pyrosequencing at specific CpG sites and LINE1 (global genome-wide DNA methylation assay) | DNA methylation | 140 women with persistent pain after breast cancer surgery | The global DNA methylation is shown to be a pain predictive biomarker, providing useful information to allocate the patients to either a “persistent pain” or “non-persistent pain” phenotype. | 31775878 | Kringel et al., 2019 [35] |
| Opioids | Tissues/Sample | Epigenetic Methods | Change | Organism | Findings | PMID | Authors |
|---|---|---|---|---|---|---|---|
| Oxycodone | Rat brains | Quantitative RT-PCR | Gene expression | Timed pregnant Sprague-Dawley rats | Exposed rat pups have lower birth weight and postnatal weight gain and greater congenital malformations. Endothelin B receptor expression is altered in the brain of oxycodone-treated rat pups indicating a possible delay in CNS(central nervous system) development. | 26676852 | Devarapalli et al., 2016 [36] |
| Methadone or buprenorphine | Cord blood or saliva | Sequencing of bisulfite-treated DNA | Determination of Percent DNA Methylation | 86 infants with Neonatal Abstinence Syndrome from mothers receiving methadone or buprenorphine during pregnancy | High methylation of three specific CpG sites of the OPRM1 promoter identified is associated with a worse outcome for Neonatal Abstinence Syndrome. | 24996986 | Wachman et al., 2014 [37] |
| Oxycodone | Brain-derived extracellular vesicle | RNA sequencing | MicroRNA expression | Male and female Sprague Dawley rats | Distinct miRNA signatures are identified in brain-derived extracellular vesicle at a key stage of brain development in offspring that were in utero and postnatal oxycodone-exposed. | 31861723 | Shahjin et al. 2019 [38] |
| Morphine | NAc | Quantitative RT-PCR | Gene expression | Female Sprague-Dawley rats | Increased expression of KOR (K opioid receptor gene) and DRD2 (Dopamine 2 receptor gene) in response to repeated administration of quinpirole (a D2/D3 receptors agonist) in the progeny of females exposed to opiates during adolescence (also observed in the F2 generations). | 23314440 | Byrnes et al., 2013 [39] |
| Morphine self-administration | NAc | RNA deep sequencing and qPCR | Gene expression | Female adolescent Sprague Dawley rats | Genes related to synaptic plasticity and the myelin basic protein are dysregulated; some effects persisted into the subsequent generation. | 27729240 | Vassoler et al., 2017 [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerra, M.C.; Dallabona, C.; Arendt-Nielsen, L. Epigenetic Alterations in Prescription Opioid Misuse: New Strategies for Precision Pain Management. Genes 2021, 12, 1226. https://doi.org/10.3390/genes12081226
Gerra MC, Dallabona C, Arendt-Nielsen L. Epigenetic Alterations in Prescription Opioid Misuse: New Strategies for Precision Pain Management. Genes. 2021; 12(8):1226. https://doi.org/10.3390/genes12081226
Chicago/Turabian StyleGerra, Maria Carla, Cristina Dallabona, and Lars Arendt-Nielsen. 2021. "Epigenetic Alterations in Prescription Opioid Misuse: New Strategies for Precision Pain Management" Genes 12, no. 8: 1226. https://doi.org/10.3390/genes12081226
APA StyleGerra, M. C., Dallabona, C., & Arendt-Nielsen, L. (2021). Epigenetic Alterations in Prescription Opioid Misuse: New Strategies for Precision Pain Management. Genes, 12(8), 1226. https://doi.org/10.3390/genes12081226