Hearing Function: Identification of New Candidate Genes Further Explaining the Complexity of This Sensory Ability
Abstract
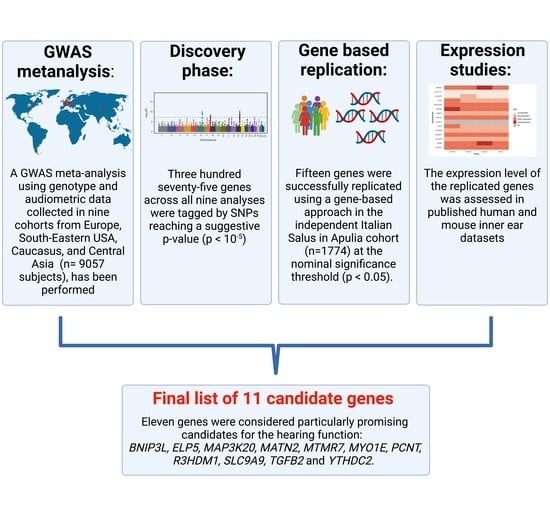
:1. Introduction
2. Materials and Methods
2.1. Involved Cohorts
2.2. Analysed Phenotypes
2.3. Genotyping, Quality Control, and Imputation
2.4. GWAS Analysis and Meta-Analysis
2.5. Replication of the Detected Genes in an Independent Cohort
2.6. Gene Expression in the Inner Ear
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fuchs, J.C.; Tucker, A.S. Development and Integration of the Ear. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2015; Volume 115, pp. 213–232. [Google Scholar] [CrossRef]
- Raynor, L.A.; Pankow, J.S.; Miller, M.B.; Huang, G.-H.; Dalton, D.; Klein, R.; Klein, B.E.K.; Cruickshanks, K.J. Familial Aggregation of Age-Related Hearing Loss in an Epidemiological Study of Older Adults. Am. J. Audiol. 2009, 18, 114–118. [Google Scholar] [CrossRef] [Green Version]
- Kvestad, E.; Czajkowski, N.; Krog, N.H.; Engdahl, B.; Tambs, K. Heritability of Hearing Loss. Epidemiology 2012, 23, 328–331. [Google Scholar] [CrossRef]
- Bogo, R.; Farah, A.; Johnson, A.-C.; Karlsson, K.K.; Pedersen, N.L.; Svartengren, M.; Skjönsberg, Å. The Role of Genetic Factors for Hearing Deterioration Across 20 Years: A Twin Study. Gerona 2015, 70, 647–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Momi, S.K.; Wolber, L.E.; Fabiane, S.M.; MacGregor, A.J.; Williams, F.M.K. Genetic and Environmental Factors in Age-Related Hearing Impairment. Twin Res. Hum. Genet. 2015, 18, 383–392. [Google Scholar] [CrossRef] [Green Version]
- Duan, H.; Zhang, D.; Liang, Y.; Xu, C.; Wu, Y.; Tian, X.; Pang, Z.; Tan, Q.; Li, S.; Qiu, C. Heritability of Age-Related Hearing Loss in Middle-Aged and Elderly Chinese: A Population-Based Twin Study. Ear Hear. 2019, 40, 253–259. [Google Scholar] [CrossRef]
- Girotto, G.; Vuckovic, D.; Buniello, A.; Lorente-Cánovas, B.; Lewis, M.; Gasparini, P.; Steel, K.P. Expression and Replication Studies to Identify New Candidate Genes Involved in Normal Hearing Function. PLoS ONE 2014, 9, e85352. [Google Scholar] [CrossRef] [PubMed]
- Wolber, L.E.; Girotto, G.; Buniello, A.; Vuckovic, D.; Pirastu, N.; Lorente-Cánovas, B.; Rudan, I.; Hayward, C.; Polasek, O.; Ciullo, M.; et al. Salt-Inducible Kinase 3, SIK3, Is a New Gene Associated with Hearing. Hum. Mol. Genet. 2014, 23, 6407–6418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuckovic, D.; Dawson, S.; Scheffer, D.I.; Rantanen, T.; Morgan, A.; Di Stazio, M.; Vozzi, D.; Nutile, T.; Concas, M.P.; Biino, G.; et al. Genome-Wide Association Analysis on Normal Hearing Function Identifies PCDH20 and SLC28A3 as Candidates for Hearing Function and Loss. Hum. Mol. Genet. 2015, 24, 5655–5664. [Google Scholar] [CrossRef] [Green Version]
- Nagtegaal, A.P.; Broer, L.; Zilhao, N.R.; Jakobsdottir, J.; Bishop, C.E.; Brumat, M.; Christiansen, M.W.; Cocca, M.; Gao, Y.; Heard-Costa, N.L.; et al. Genome-Wide Association Meta-Analysis Identifies Five Novel Loci for Age-Related Hearing Impairment. Sci. Rep. 2019, 9, 15192. [Google Scholar] [CrossRef] [PubMed]
- Cocca, M.; Barbieri, C.; Concas, M.P.; Robino, A.; Brumat, M.; Gandin, I.; Trudu, M.; Sala, C.F.; Vuckovic, D.; Girotto, G.; et al. A Bird’s-Eye View of Italian Genomic Variation through Whole-Genome Sequencing. Eur. J. Hum. Genet. 2020, 28, 435–444. [Google Scholar] [CrossRef]
- Mezzavilla, M.; Vozzi, D.; Pirastu, N.; Girotto, G.; d’Adamo, P.; Gasparini, P.; Colonna, V. Genetic Landscape of Populations along the Silk Road: Admixture and Migration Patterns. BMC Genet. 2014, 15, 131. [Google Scholar] [CrossRef] [Green Version]
- Van Eyken, E.; Van Laer, L.; Fransen, E.; Topsakal, V.; Lemkens, N.; Laureys, W.; Nelissen, N.; Vandevelde, A.; Wienker, T.; Van De Heyning, P.; et al. KCNQ4: A Gene for Age-Related Hearing Impairment? Hum. Mutat. 2006, 27, 1007–1016. [Google Scholar] [CrossRef]
- Ikram, M.A.; Brusselle, G.G.O.; Murad, S.D.; van Duijn, C.M.; Franco, O.H.; Goedegebure, A.; Klaver, C.C.W.; Nijsten, T.E.C.; Peeters, R.P.; Stricker, B.H.; et al. The Rotterdam Study: 2018 Update on Objectives, Design and Main Results. Eur. J. Epidemiol. 2017, 32, 807–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigters, S.C.; van der Schroeff, M.P.; Papageorgiou, G.; de Jong, R.J.B.; Goedegebure, A. Progression of Hearing Loss in the Aging Population: Repeated Auditory Measurements in the Rotterdam Study. Audiol. Neurotol. 2018, 23, 290–297. [Google Scholar] [CrossRef]
- Tsao, C.W.; Vasan, R.S. Cohort Profile: The Framingham Heart Study (FHS): Overview of Milestones in Cardiovascular Epidemiology. Int. J. Epidemiol. 2015, 44, 1800–1813. [Google Scholar] [CrossRef] [Green Version]
- Sardone, R.; Battista, P.; Donghia, R.; Lozupone, M.; Tortelli, R.; Guerra, V.; Grasso, A.; Griseta, C.; Castellana, F.; Zupo, R.; et al. Age-Related Central Auditory Processing Disorder, MCI, and Dementia in an Older Population of Southern Italy. Otolaryngol. Head Neck Surg. 2020, 163, 348–355. [Google Scholar] [CrossRef]
- Girotto, G.; Pirastu, N.; Sorice, R.; Biino, G.; Campbell, H.; d’Adamo, A.P.; Hastie, N.D.; Nutile, T.; Polasek, O.; Portas, L.; et al. Hearing Function and Thresholds: A Genome-Wide Association Study in European Isolated Populations Identifies New Loci and Pathways. J. Med. Genet. 2011, 48, 369–374. [Google Scholar] [CrossRef] [Green Version]
- The 1000 Genomes Project Consortium. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Haplotype Reference Consortium. A Reference Panel of 64,976 Haplotypes for Genotype Imputation. Nat. Genet. 2016, 48, 1279–1283. [Google Scholar] [CrossRef] [Green Version]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; van Duijn, C.M. GenABEL: An R Library for Genome-Wide Association Analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svishcheva, G.R.; Axenovich, T.I.; Belonogova, N.M.; van Duijn, C.M.; Aulchenko, Y.S. Rapid Variance Components–Based Method for Whole-Genome Association Analysis. Nat. Genet. 2012, 44, 1166–1170. [Google Scholar] [CrossRef]
- Zhan, X.; Hu, Y.; Li, B.; Abecasis, G.R.; Liu, D.J. RVTESTS: An Efficient and Comprehensive Tool for Rare Variant Association Analysis Using Sequence Data: Table 1. Bioinformatics 2016, 32, 1423–1426. [Google Scholar] [CrossRef] [Green Version]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and Efficient Meta-Analysis of Genomewide Association Scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Stephens, M. Genome-Wide Efficient Mixed-Model Analysis for Association Studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [Green Version]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional Mapping and Annotation of Genetic Associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrauwen, I.; Hasin-Brumshtein, Y.; Corneveaux, J.J.; Ohmen, J.; White, C.; Allen, A.N.; Lusis, A.J.; Van Camp, G.; Huentelman, M.J.; Friedman, R.A. A Comprehensive Catalogue of the Coding and Non-Coding Transcripts of the Human Inner Ear. Hear. Res. 2016, 333, 266–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elkon, R.; Milon, B.; Morrison, L.; Shah, M.; Vijayakumar, S.; Racherla, M.; Leitch, C.C.; Silipino, L.; Hadi, S.; Weiss-Gayet, M.; et al. RFX Transcription Factors Are Essential for Hearing in Mice. Nat. Commun. 2015, 6, 8549. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Garcia-Etxebarria, K.; Jauregi-Miguel, A.; Romero-Garmendia, I.; Plaza-Izurieta, L.; Legarda, M.; Irastorza, I.; Bilbao, J.R. Ancestry-Based Stratified Analysis of Immunochip Data Identifies Novel Associations with Celiac Disease. Eur. J. Hum. Genet. 2016, 24, 1831–1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patak, J.; Faraone, S.V.; Zhang-James, Y. Sodium Hydrogen Exchanger 9 NHE9 (SLC9A9) and Its Emerging Roles in Neuropsychiatric Comorbidity. Am. J. Med. Genet. 2020, 183, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang-James, Y.; Vaudel, M.; Mjaavatten, O.; Berven, F.S.; Haavik, J.; Faraone, S.V. Effect of Disease-Associated SLC9A9 Mutations on Protein–Protein Interaction Networks: Implications for Molecular Mechanisms for ADHD and Autism. Atten. Deficit Hyperact. Disord. 2019, 11, 91–105. [Google Scholar] [CrossRef]
- Kucharava, K.; Brand, Y.; Albano, G.; Sekulic-Jablanovic, M.; Glutz, A.; Xian, X.; Herz, J.; Bodmer, D.; Fuster, D.G.; Petkovic, V. Sodium-Hydrogen Exchanger 6 (NHE6) Deficiency Leads to Hearing Loss, via Reduced Endosomal Signalling through the BDNF/Trk Pathway. Sci. Rep. 2020, 10, 3609. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ma, C.; Wang, K.; Li, X.; Jian, X.; Zhang, H.; Yuan, J.; Yin, J.; Chen, J.; Shi, Y. Identification of Rare and Common Variants in BNIP3L: A Schizophrenia Susceptibility Gene. Hum. Genom. 2020, 14, 16. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Choo, O.-S.; Lee, J.-S.; Jang, J.H.; Woo, H.G.; Choung, Y.-H. BCL2 Interacting Protein 3-like/NIX-Mediated Mitophagy Plays an Important Role in the Process of Age-Related Hearing Loss. Neuroscience 2021, 455, 39–51. [Google Scholar] [CrossRef]
- Youn, C.K.; Jun, Y.; Jo, E.-R.; Cho, S.I. Age-Related Hearing Loss in C57BL/6J Mice Is Associated with Mitophagy Impairment in the Central Auditory System. Int. J. Mol. Sci. 2020, 21, 7202. [Google Scholar] [CrossRef]
- Liu, T.-C.; Huang, C.-J.; Chu, Y.-C.; Wei, C.-C.; Chou, C.-C.; Chou, M.-Y.; Chou, C.-K.; Yang, J.-J. Cloning and Expression of ZAK, a Mixed Lineage Kinase-like Protein Containing a Leucine-Zipper and a Sterile-Alpha Motif. Biochem. Biophys. Res. Commun. 2000, 274, 811–816. [Google Scholar] [CrossRef]
- Cho, Y.-Y.; Bode, A.M.; Mizuno, H.; Choi, B.Y.; Choi, H.S.; Dong, Z. A Novel Role for Mixed-Lineage Kinase-Like Mitogen-Activated Protein Triple Kinase α in Neoplastic Cell Transformation and Tumor Development. Cancer Res. 2004, 64, 3855–3864. [Google Scholar] [CrossRef] [Green Version]
- Muratoglu, S.; Krysan, K.; Balázs, M.; Sheng, H.; Zákány, R.; Módis, L.; Kiss, I.; Deák, F. Primary Structure of Human Matrilin-2, Chromosome Location of the MATN2 Gene and Conservation of an AT-AC Intron in Matrilin Genes. Cytogenet. Genome Res. 2000, 90, 323–327. [Google Scholar] [CrossRef]
- Korpos, É.; Deák, F.; Kiss, I. Matrilin-2, an Extracellular Adaptor Protein, Is Needed for the Regeneration of Muscle, Nerve and Other Tissues. Neural Regen. Res. 2015, 10, 866. [Google Scholar] [CrossRef]
- Cirilo, J.A.; Gunther, L.K.; Yengo, C.M. Functional Role of Class III Myosins in Hair Cells. Front. Cell Dev. Biol. 2021, 9, 643856. [Google Scholar] [CrossRef]
- Mele, C.; Iatropoulos, P.; Donadelli, R.; Calabria, A.; Maranta, R.; Cassis, P.; Buelli, S.; Tomasoni, S.; Piras, R.; Krendel, M.; et al. MYO1E Mutations and Childhood Familial Focal Segmental Glomerulosclerosis. N. Engl. J. Med. 2011, 365, 295–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumont, R.A.; Zhao, Y.; Holt, J.R.; Bähler, M.; Gillespie, P.G. Myosin-I Isozymes in Neonatal Rodent Auditory and Vestibular Epithelia. J. Assoc. Res. Otolaryngol. 2002, 3, 375–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murillo-Cuesta, S.; Rodrà guez-de la Rosa, L.; Contreras, J.; Celaya, A.M.; Camarero, G.; Rivera, T.; Varela-Nieto, I. Transforming Growth Factor Î21 Inhibition Protects from Noise-Induced Hearing Loss. Front. Aging Neurosci. 2015, 7, 32. [Google Scholar] [CrossRef] [Green Version]
- Sanford, L.P.; Ormsby, I.; Gittenberger-de Groot, A.C.; Sariola, H.; Friedman, R.; Boivin, G.P.; Cardell, E.L.; Doetschman, T. TGFbeta2 Knockout Mice Have Multiple Developmental Defects That Are Non-Overlapping with Other TGFbeta Knockout Phenotypes. Development 1997, 124, 2659–2670. [Google Scholar] [CrossRef]
- Tanabe, A.; Konno, J.; Tanikawa, K.; Sahara, H. Transcriptional Machinery of TNF-α-Inducible YTH Domain Containing 2 (YTHDC2) Gene. Gene 2014, 535, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Iossifov, I.; O’Roak, B.J.; Sanders, S.J.; Ronemus, M.; Krumm, N.; Levy, D.; Stessman, H.A.; Witherspoon, K.T.; Vives, L.; Patterson, K.E.; et al. The Contribution of de Novo Coding Mutations to Autism Spectrum Disorder. Nature 2014, 515, 216–221. [Google Scholar] [CrossRef] [Green Version]
- Fanale, D.; Iovanna, J.L.; Calvo, E.L.; Berthezene, P.; Belleau, P.; Dagorn, J.C.; Bronte, G.; Cicero, G.; Bazan, V.; Rolfo, C.; et al. Germline Copy Number Variation in the YTHDC2 Gene: Does It Have a Role in Finding a Novel Potential Molecular Target Involved in Pancreatic Adenocarcinoma Susceptibility? Expert Opin. Ther. Targets 2014, 18, 841–850. [Google Scholar] [CrossRef] [Green Version]
- Dalwadi, U.; Yip, C.K. Structural Insights into the Function of Elongator. Cell. Mol. Life Sci. 2018, 75, 1613–1622. [Google Scholar] [CrossRef]
- Close, P.; Gillard, M.; Ladang, A.; Jiang, Z.; Papuga, J.; Hawkes, N.; Nguyen, L.; Chapelle, J.-P.; Bouillenne, F.; Svejstrup, J.; et al. DERP6 (ELP5) and C3ORF75 (ELP6) Regulate Tumorigenicity and Migration of Melanoma Cells as Subunits of Elongator. J. Biol. Chem. 2012, 287, 32535–32545. [Google Scholar] [CrossRef] [Green Version]
- Belalcazar, L.M.; Papandonatos, G.D.; McCaffery, J.M.; Peter, I.; Pajewski, N.M.; Erar, B.; Allred, N.D.; Balasubramanyam, A.; Bowden, D.W.; Brautbar, A.; et al. A Common Variant in the CLDN7/ELP5 Locus Predicts Adiponectin Change with Lifestyle Intervention and Improved Fitness in Obese Individuals with Diabetes. Physiol. Genom. 2015, 47, 215–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Guo, Y.; Liu, X.; Zhang, R.; Dong, J.; Deng, H.; He, F.; Che, F.; Liu, S.; Yi, M. Family-Based Analysis Combined with Case–Controls Study Implicate Roles of PCNT in Tourette Syndrome. Neuropsychiatr. Dis. Treat. 2020, 16, 349–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eggermont, J.J. Cochlea and auditory nerve. Handb. Clin. Neurol. 2019, 160, 437–449. [Google Scholar] [CrossRef] [PubMed]



| Discovery GWAS Meta-Analysis | Replication Analysis | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Trait | SNP | Chr | Position | Effect | Alleles | Freq | N | Beta | StdErr | p-Value | Direction | p-Value PTAM | p-Value PTAH |
| BNIP3L | 8 kHz | rs17402216 | 8 | 26325225 | intron | A/C | 0.944 | 9003 | 0.043 | 0.008 | 4.69 × 10−7 | ++-+-++++ | 5.99 × 10−3 | 2.56 × 10−3 |
| CCDC177 | PTAM | rs2025133 | 14 | 70058292 | upstream gene | A/G | 0.829 | 8989 | −0.021 | 0.005 | 3.87 × 10−6 | --------- | 2.49 × 10−1 | 1.38 × 10−2 |
| DLGAP2 | 500 Hz | rs181305232 | 8 | 1539701 | intron | T/C | 0.034 | 6152 | −0.082 | 0.018 | 5.68 × 10−6 | ---??---? | 7.03 × 10−1 | 4.57 × 10−3 |
| ELP5 | 4 kHz | rs2106842 | 17 | 7162451 | intron | A/G | 0.646 | 5729 | −0.022 | 0.005 | 2.84 × 10−6 | --++---?? | 3.58 × 10−2 | 6.69 × 10−1 |
| PTAH | 5729 | −0.015 | 0.003 | 6.23 × 10−6 | --+----?? | |||||||||
| MAP3K20 | PTAM | rs113132813 | 2 | 173956255 | intron | A/G | 0.021 | 5801 | −0.075 | 0.016 | 1.49 × 10−6 | ?-+??---- | 4.42 × 10−2 | 6.97 × 10−1 |
| MATN2 | 1 kHz | rs11996075 | 8 | 98910287 | intron | T/C | 0.118 | 7932 | 0.034 | 0.007 | 4.83 × 10−6 | ++++?+++- | 1.28 × 10−1 | 4.22 × 10−2 |
| MTMR7 | 1 kHz | rs12155974 | 8 | 17299124 | upstream gene | T/C | 0.911 | 7932 | −0.050 | 0.010 | 2.35 × 10−7 | ----?---- | 7.04 × 10−3 | 8.09 × 10−4 |
| MYO1E | 2 kHz | rs67412566 | 15 | 59523608 | intron | A/T | 0.833 | 6820 | 0.048 | 0.011 | 7.66 × 10−6 | ++++?+++? | 4.19 × 10−2 | 9.20 × 10−1 |
| OOSP1 | PTAH | rs111389524 | 11 | 59757661 | downstream gene | A/G | 0.036 | 7895 | −0.044 | 0.010 | 7.14 × 10−6 | ----?---- | 4.21 × 10−4 | 1.55 × 10−2 |
| PCNT | 8 kHz | rs11909986 | 21 | 47780161 | intron | A/G | 0.332 | 8348 | −0.018 | 0.004 | 7.86 × 10−6 | ---?----- | 1.87 × 10−1 | 3.02 × 10−2 |
| PTPRN2 | 1 kHz | rs11514653 | 7 | 158356977 | intron | C/G | 0.022 | 6954 | 0.091 | 0.019 | 2.78 × 10−6 | ++??++++? | 4.20 × 10−1 | 4.78 × 10−2 |
| R3HDM1 | 500 Hz | rs7560535 | 2 | 136412472 | intron | A/G | 0.175 | 6431 | 0.032 | 0.007 | 7.39 × 10−6 | ?+++?++++ | 9.25 × 10−3 | 2.41 × 10−1 |
| SLC9A9 | 250 Hz | rs76168782 | 3 | 143270684 | intron | T/G | 0.946 | 7244 | −0.054 | 0.012 | 6.86 × 10−6 | ---??---- | 2.47 × 10−3 | 8.60 × 10−1 |
| PTAL | 7264 | −0.046 | 0.010 | 3.27 × 10−6 | ---??---- | |||||||||
| TGFB2 | 8 kHz | rs149269977 | 1 | 218546474 | intron | T/C | 0.026 | 7874 | −0.071 | 0.016 | 7.86 × 10−6 | ---+----? | 2.50 × 10−2 | 4.06 × 10−1 |
| YTHDC2 | 8 kHz | rs77567880 | 5 | 113145730 | downstream gene | A/G | 0.015 | 6055 | −0.102 | 0.022 | 4.36 × 10−6 | ??--?--+- | 3.14 × 10−1 | 4.84 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Concas, M.P.; Morgan, A.; Serra, F.; Nagtegaal, A.P.; Oosterloo, B.C.; Seshadri, S.; Heard-Costa, N.; Van Camp, G.; Fransen, E.; Francescatto, M.; et al. Hearing Function: Identification of New Candidate Genes Further Explaining the Complexity of This Sensory Ability. Genes 2021, 12, 1228. https://doi.org/10.3390/genes12081228
Concas MP, Morgan A, Serra F, Nagtegaal AP, Oosterloo BC, Seshadri S, Heard-Costa N, Van Camp G, Fransen E, Francescatto M, et al. Hearing Function: Identification of New Candidate Genes Further Explaining the Complexity of This Sensory Ability. Genes. 2021; 12(8):1228. https://doi.org/10.3390/genes12081228
Chicago/Turabian StyleConcas, Maria Pina, Anna Morgan, Fabrizio Serra, Andries Paul Nagtegaal, Berthe C. Oosterloo, Sudha Seshadri, Nancy Heard-Costa, Guy Van Camp, Erik Fransen, Margherita Francescatto, and et al. 2021. "Hearing Function: Identification of New Candidate Genes Further Explaining the Complexity of This Sensory Ability" Genes 12, no. 8: 1228. https://doi.org/10.3390/genes12081228
APA StyleConcas, M. P., Morgan, A., Serra, F., Nagtegaal, A. P., Oosterloo, B. C., Seshadri, S., Heard-Costa, N., Van Camp, G., Fransen, E., Francescatto, M., Logroscino, G., Sardone, R., Quaranta, N., Gasparini, P., & Girotto, G. (2021). Hearing Function: Identification of New Candidate Genes Further Explaining the Complexity of This Sensory Ability. Genes, 12(8), 1228. https://doi.org/10.3390/genes12081228