Annotation and Analysis of 3902 Odorant Receptor Protein Sequences from 21 Insect Species Provide Insights into the Evolution of Odorant Receptor Gene Families in Solitary and Social Insects
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Data Retrieval, Curation and Alignment
2.2. Clustering of OR Proteins
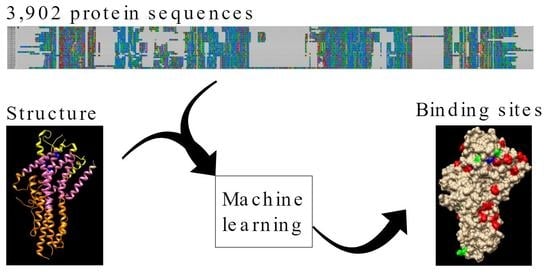
2.3. Machine Learning Approach
2.4. Computation of Sequence Conservation
3. Results
3.1. Collection and Curation of Insect Odorant Receptor Proteins
3.2. The Taxonomic Distribution of ORs in Clusters Shows Taxa-Specific Expansions
3.3. Prediction of OR Amino Acid Residues Important for Chemical Binding
3.4. Relative Conservation of Predictive Residues
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brand, P.; Robertson, H.M.; Lin, W.; Pothula, R.; Klingeman, W.E.; Jurat-Fuentes, J.L.; Johnson, B.R. The origin of the odorant receptor gene family in insects. eLife 2018, 7, e38340. [Google Scholar] [CrossRef] [PubMed]
- Brand, P.; Hinojosa-Díaz, I.A.; Ayala, R.; Daigle, M.; Yurrita Obiols, C.L.; Eltz, T.; Ramírez, S.R. The evolution of sexual signaling is linked to odorant receptor tuning in perfume-collecting orchid bees. Nat. Commun. 2020, 11, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, M.; Du, L.; Chen, Q.; Feng, Y.; Zhang, J.; Zhang, X.; Tian, K.; Cao, S.; Huang, T.; Jacquin-Joly, E.; et al. Odorant Receptors for Detecting Flowering Plant Cues Are Functionally Conserved across Moths and Butterflies. Mol. Biol. Evol. 2021, 38, 1413–1427. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, F.; Borrero-Echeverry, F.; Jósvai, J.K.; Strandh, M.; Unelius, C.R.; Tóth, M.; Witzgall, P.; Bengtsson, M.; Walker, W.B., III. Odorant receptor phylogeny confirms conserved channels for sex pheromone and host plant signals in tortricid moths. Ecol. Evol. 2020, 10, 7334–7348. [Google Scholar] [CrossRef]
- Legan, A.W.; Jernigan, C.M.; Miller, S.E.; Fuchs, M.F.; Sheehan, M.J. Expansion and Accelerated Evolution of 9-Exon Odorant Receptors in Polistes Paper Wasps. Mol. Biol. Evol. 2021, 38, 3832–3846. [Google Scholar] [CrossRef]
- Smith, C.D.; Zimin, A.; Holt, C.; Abouheif, E.; Benton, R.; Cash, E.; Croset, V.; Currie, C.R.; Elhaik, E.; Elsik, C.G.; et al. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc. Natl. Acad. Sci. USA 2011, 108, 5673–5678. [Google Scholar] [CrossRef] [Green Version]
- Kapheim, K.M.; Pan, H.; Li, C.; Salzberg, S.L.; Puiu, D.; Magoc, T.; Robertson, H.M.; Hudson, M.E.; Venkat, A.; Fischman, B.J.; et al. Social evolution. Genomic signatures of evolutionary transitions from solitary to group living. Science 2015, 348, 1139–1143. [Google Scholar] [CrossRef] [Green Version]
- Hölldobler, B.; Wilson, E.O. The Ants, Harvard University Press; The Belknap Press of Harvard University: Cambridge, MA, USA, 1990. [Google Scholar]
- Wilson, E.O. Chemical Communication in the Social Insects. Science 1965, 149, 1064–1071. [Google Scholar] [CrossRef]
- Taylor, B.A.; Cini, A.; Wyatt, C.D.R.; Reuter, M.; Sumner, S. The molecular basis of socially mediated phenotypic plasticity in a eusocial paper wasp. Nat. Commun. 2021, 12, 775. [Google Scholar] [CrossRef]
- Leonhardt, S.D.; Menzel, F.; Nehring, V.; Schmitt, T. Ecology and Evolution of Communication in Social Insects. Cell 2016, 164, 1277–1287. [Google Scholar] [CrossRef] [Green Version]
- McKenzie, S.K.; Kronauer, D.J.C. The genomic architecture and molecular evolution of ant odorant receptors. Genome Res. 2018, 28, 1757–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trible, W.; Olivos-Cisneros, L.; McKenzie, S.K.; Saragosti, J.; Chang, N.C.; Matthews, B.J.; Oxley, P.R.; Kronauer, D.J. Orco Mutagenesis Causes Loss of Antennal Lobe Glomeruli and Impaired Social Behavior in Ants. Cell 2017, 170, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Opachaloemphan, C.; Mancini, G.; Yang, H.; Gallitto, M.; Mlejnek, J.; Leibholz, A.; Haight, K.; Ghaninia, M.; Huo, L.; et al. An Engineered orco Mutation Produces Aberrant Social Behavior and Defective Neural Development in Ants. Cell 2017, 170, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Jongepier, E.; Séguret, A.; Labutin, A.; Feldmeyer, B.; Gstöttl, C.; Foitzik, S.; Heinze, J.; Bornberg-Bauer, E. Convergent loss of chemoreceptors across independent origins of slave-making in ants. Mol. Biol. Evol. 2021, 20, msab305. [Google Scholar] [CrossRef] [PubMed]
- Schrader, L.; Pan, H.; Bollazzi, M.; Schiøtt, M.; Larabee, F.J.; Bi, X.; Deng, Y.; Zhang, G.; Boomsma, J.J.; Rabeling, C. Relaxed selection underlies genome erosion in socially parasitic ant species. Nat. Commun. 2021, 12, 2918. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Slone, J.D.; Rokas, A.; Berger, S.L.; Liebig, J.; Ray, A.; Reinberg, D.; Zwiebel, L.J. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLoS Genet. 2012, 8, e1002930. [Google Scholar] [CrossRef] [Green Version]
- Engsontia, P.; Sangket, U.; Robertson, H.M.; Satasook, C. Diversification of the ant odorant receptor gene family and positive selection on candidate cuticular hydrocarbon receptors. BMC Res. Notes 2015, 8, 380. [Google Scholar] [CrossRef] [Green Version]
- McKenzie, S.K.; Fetter-Pruneda, I.; Ruta, V.; Kronauer, D.J.C. Transcriptomics and neuroanatomy of the clonal raider ant implicate an expanded clade of odorant receptors in chemical communication. Proc. Natl. Acad. Sci. USA 2016, 113, 14091–14096. [Google Scholar] [CrossRef] [Green Version]
- Saad, R.; Cohanim, A.B.; Kosloff, M.; Privman, E. Neofunctionalization in Ligand Binding Sites of Ant Olfactory Receptors. Genome Biol. Evol. 2018, 10, 2490–2500. [Google Scholar] [CrossRef] [Green Version]
- McKenzie, S.K.; Winston, M.E.; Grewe, F.; Vargas Asensio, G.; Rodríguez-Hernández, N.; Rubin, B.E.; Murillo-Cruz, C.; von Beeren, C.; Moreau, C.S.; Suen, G.; et al. The genomic basis of army ant chemosensory adaptations. Mol. Ecol. 2021, 24, 5527–6641. [Google Scholar] [CrossRef]
- Pask, G.M.; Slone, J.D.; Millar, J.G.; Das, P.; Moreira, J.A.; Zhou, X.; Bello, J.; Berger, S.L.; Bonasio, R.; Desplan, C.; et al. Specialized odorant receptors in social insects that detect cuticular hydrocarbon cues and candidate pheromones. Nat. Commun. 2017, 8, 297. [Google Scholar] [CrossRef] [PubMed]
- Slone, J.D.; Pask, G.M.; Ferguson, S.T.; Millar, J.G.; Berger, S.L.; Reinberg, D.; Liebig, J.; Ray, A.; Zwiebel, L.J. Functional characterization of odorant receptors in the ponerine ant, Harpegnathos saltator. Proc. Natl. Acad. Sci. USA 2017, 114, 8586–8591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malnic, B.; Hirono, J.; Sato, T.; Buck, L.B. Combinatorial receptor codes for odors. Cell 1999, 96, 713–723. [Google Scholar] [CrossRef] [Green Version]
- Wicher, D. Tuning Insect Odorant Receptors. Front. Cell. Neurosci. 2018, 12, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carey, A.F.; Wang, G.; Su, C.-Y.; Zwiebel, L.J.; Carlson, J.R. Odorant reception in the malaria mosquito Anopheles gambiae. Nature 2010, 464, 66–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Münch, D.; Galizia, C.G. DoOR 2.0—Comprehensive Mapping of Drosophila melanogaster Odorant Responses. Sci. Rep. 2016, 6, 21841. [Google Scholar] [CrossRef] [Green Version]
- Kepchia, D.; Xu, P.; Terryn, R.; Castro, A.; Schürer, S.C.; Leal, W.S.; Luetje, C.W. Use of machine learning to identify novel, behaviorally active antagonists of the insect odorant receptor co-receptor (Orco) subunit. Sci. Rep. 2019, 9, 4055. [Google Scholar] [CrossRef] [Green Version]
- Caballero-Vidal, G.; Bouysset, C.; Grunig, H.; Fiorucci, S.; Montagné, N.; Golebiowski, J.; Jacquin-Joly, E. Machine learning decodes chemical features to identify novel agonists of a moth odorant receptor. Sci. Rep. 2020, 10, 1655. [Google Scholar] [CrossRef]
- Butterwick, J.A.; Del Mármol, J.; Kim, K.H.; Kahlson, M.A.; Rogow, J.A.; Walz, T.; Ruta, V. Cryo-EM structure of the insect olfactory receptor Orco. Nature 2018, 560, 447–452. [Google Scholar] [CrossRef]
- Robertson, H.M.; Warr, C.G.; Carlson, J.R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2003, 100, 14537–14542. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.R.; Smith, C.D.; Robertson, H.M.; Helmkampf, M.; Zimin, A.; Yandell, M.; Holt, C.; Hu, H.; Abouheif, E.; Benton, R.; et al. Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc. Natl. Acad. Sci. USA 2011, 108, 5668–5672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, H.M.; Baits, R.L.; Walden, K.K.O.; Wada-Katsumata, A.; Schal, C. Enormous expansion of the chemosensory gene repertoire in the omnivorous German cockroach Blattela germanica. J. Exp. Zool. B Mol. Dev. Evol. 2018, 330, 265–278. [Google Scholar] [CrossRef] [Green Version]
- Ioannidis, P.; Simao, F.A.; Waterhouse, R.M.; Manni, M.; Seppey, M.; Robertson, H.M.; Misof, B.; Niehuis, O.; Zdobnov, E.M. Genomic Features of the Damselfly Calopteryx splendens Representing a Sister Clade to Most Insect orders. Genome Biol. Evol. 2017, 9, 415–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadd, B.M.; Barribeau, S.M.; Bloch, G.; De Graaf, D.C.; Dearden, P.; Elsik, C.G.; Gadau, J.; Grimmelikhuijzen, C.J.; Hasselmann, M.; Lozier, J.D.; et al. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 2015, 16, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tribolium Genome Sequencing Consortium; Richards, S.; Gibbs, R.A.; Weinstock, G.M.; Brown, S.J.; Denell, R.; Beeman, R.W.; Gibbs, R.; Bucher, G.; Friedrich, M.; et al. The genome of the model beetle and pest Tribolium castaneum. Nature 2008, 452, 949–955. [Google Scholar] [PubMed] [Green Version]
- Almudi, I.; Vizueta, J.; Wyatt, C.D.; de Mendoza, A.; Marlétaz, F.; Firbas, P.N.; Feuda, R.; Masiero, G.; Medina, P.; Alcaina-Caro, A.; et al. Genomic adaptations to aquatic and aerial life in mayflies and the origin of insect wings. Nat. Commun. 2020, 11, 2631. [Google Scholar] [CrossRef]
- Koenig, C.; Hirsh, A.; Bucks, S.; Klinner, C.; Vogel, H.; Shukla, A.; Mansfield, J.H.; Morton, B.; Hansson, B.S.; Grosse-Wilde, E.; et al. A reference gene set for chemosensory receptor genes of Manduca sexta. Insect Biochem. Mol. Biol. 2015, 66, 51–63. [Google Scholar] [CrossRef]
- Kirkness, E.F.; Haas, B.J.; Sun, W.; Braig, H.R.; Perotti, M.A.; Clark, J.M.; Lee, S.H.; Robertson, H.M.; Kennedy, R.C.; Elhaik, E.; et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc. Natl. Acad. Sci. USA 2010, 107, 12168–12173. [Google Scholar] [CrossRef] [Green Version]
- Robertson, H.M.; Robertson, E.C.N.; Walden, K.K.O.; Enders, L.S.; Miller, N.J. The chemoreceptors and odorant binding proteins of the soybean and pea aphids. Insect Biochem. Mol. Biol. 2019, 105, 69–78. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Borowiec, M.L.; Moreau, C.S.; Rabeling, C. Ants: Phylogeny and Classification. In Encyclopedia of Social Insects; Starr, C., Ed.; Springer: Cham, Switzerland, 2020. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, K.; Pellegrino, M.; Nakagawa, T.; Nakagawa, T.; Vosshall, L.B.; Touhara, K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 2008, 452, 1002–1006. [Google Scholar] [CrossRef] [PubMed]
- Wicher, D.; Schäfer, R.; Bauernfeind, R.; Stensmyr, M.C.; Heller, R.; Heinemann, S.H.; Hansson, B.S. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 2008, 452, 1007–1011. [Google Scholar] [CrossRef]
- Smadja, C.; Shi, P.; Butlin, R.K.; Robertson, H.M. Large gene family expansions and adaptive evolution for odorant and gustatory receptors in the pea aphid, Acyrthosiphon pisum. Mol. Biol. Evol. 2009, 26, 2073–2086. [Google Scholar] [CrossRef] [Green Version]
- Del Mármol, J.; Yedlin, M.A.; Ruta, V. The structural basis of odorant recognition in insect olfactory receptors. Nature 2021, 597, 126–131. [Google Scholar] [CrossRef]
- Nichols, A.S.; Luetje, C.W. Transmembrane segment 3 of Drosophila melanogaster odorant receptor subunit 85b contributes to ligand-receptor interactions. J. Biol. Chem. 2010, 285, 11854–11862. [Google Scholar] [CrossRef] [Green Version]
- Rahman, S.; Luetje, C.W. Mutant cycle analysis identifies a ligand interaction site in an odorant receptor of the malaria vector Anopheles gambiae. J. Biol. Chem. 2017, 292, 18916–18923. [Google Scholar] [CrossRef] [Green Version]
- Leary, G.P.; Allen, J.E.; Bunger, P.L.; Luginbill, J.B.; Linn, C.E.; Macallister, I.E.; Kavanaugh, M.P.; Wanner, K.W. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. USA 2012, 109, 14081–14086. [Google Scholar] [CrossRef] [Green Version]
- Domman, D.; Collingro, A.; Lagkouvardos, I.; Gehre, L.; Weinmaier, T.; Rattei, T.; Subtil, A.; Horn, M. Massive expansion of Ubiquitination-related gene families within the Chlamydiae. Mol. Biol. Evol. 2014, 31, 2890–2904. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Ma, H.; Nei, M.; Kong, H. Evolution of F-box genes in plants: Different modes of sequence divergence and their relationships with functional diversification. Proc. Natl. Acad. Sci. USA 2009, 106, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Species | Tax ID | Taxonomy (Order > Suborder > Family) | Raw Number OR | Curated Number OR |
|---|---|---|---|---|
| Calopteryx splendens | 52612 | Odonata > Zygoptera > Calopterygidae | 5 | 5 |
| Cloeon dipterum | 197152 | Ephemeroptera > Pisciforma > Baetidae | 50 | 24 |
| Blattella germanica | 6973 | Blattodea > - > Ectobiidae | 135 | 89 |
| Aphis glycines | 307491 | Hemiptera > Sternorrhyncha > Aphididae | 47 | 42 |
| Acyrthosiphon pisum | 7029 | Hemiptera > Sternorrhyncha > Aphididae | 87 | 67 |
| Pediculus humanus | 121224 | Phthiraptera > Anoplura > Pediculidae | 13 | 10 |
| Tribolium castaneum | 7070 | Coleoptera > Polyphaga > Tenebrionidae | 338 | 253 |
| Manduca sexta | 7130 | Lepidoptera > Glossata > Sphingidae | 74 | 50 |
| Anopheles gambiae | 7165 | Diptera > Nematocera > Culicidae | 79 | 73 |
| Drosophila melanogaster | 7227 | Diptera > Brachycera > Drosophilidae | 61 | 61 |
| Nasonia vitripennis | 7425 | Hymenoptera > Apocrita > Pteromalidae | 211 | 199 |
| Bombus terrestris | 30195 | Hymenoptera > Apocrita > Apidae | 165 | 149 |
| Apis mellifera | 7460 | Hymenoptera > Apocrita > Apidae | 160 | 153 |
| Harpegnathos saltator | 610380 | Hymenoptera > Apocrita > Formicidae | 377 | 360 |
| Ooceraea biroi | 2015173 | Hymenoptera > Apocrita > Formicidae | 574 | 501 |
| Linepithema humile | 83485 | Hymenoptera > Apocrita > Formicidae | 367 | 323 |
| Camponotus floridanus | 104421 | Hymenoptera > Apocrita > Formicidae | 407 | 376 |
| Solenopsis invicta | 13686 | Hymenoptera > Apocrita > Formicidae | 396 | 287 |
| Pogonomyrmex barbatus | 144034 | Hymenoptera > Apocrita > Formicidae | 293 | 293 |
| Acromyrmex echinatior | 103372 | Hymenoptera > Apocrita > Formicidae | 435 | 306 |
| Atta cephalotes | 12957 | Hymenoptera > Apocrita > Formicidae | 434 | 281 |
| Code | Taxon |
|---|---|
| (A) | Insecta |
| (B) | Palaeoptera |
| (B1) | C. splendens |
| (B2) | C. dipterum |
| (C) | Neoptera |
| (D) | B. germanica |
| (E) | Paraneoptera |
| (F) | Hemiptera |
| (F1) | A. glycines |
| (F2) | A. pisum |
| (G) | P. humanus |
| (H) | Endopterygota |
| (I) | T. castaneum |
| (J) | M. sexta |
| (K) | Diptera |
| (K1) | A. gambiae |
| (K2) | D. melanogaster |
| (L) | Hymenoptera |
| (M) | N. vitripennis |
| (N) | Aculeata |
| (O) | Apoidea |
| (O1) | B. terrestris |
| (O2) | A. mellifera |
| (P) | Formicoidea |
| (Q) | H. saltator |
| (R) | Formicoids |
| (S) | O. biroi |
| (T) | Formicoids − O. biroi |
| (U) | L. humile |
| (V) | Myrmicinae + C. floridanus |
| (W) | C. floridanus |
| (X) | Myrmicinae |
| (X1) | S. invicta |
| (X2) | P. barbatus |
| (Y) | Attini |
| (Y1) | A. echinatior |
| (Y2) | A. cephalotes |
| Dataset | Times Predictive | Alignment Position | D. melanogaster Orco Position | A. bakeri Orco Position | Amino Acid (D.m./A.b.) |
|---|---|---|---|---|---|
| 1 | 8 | 1472 | 207 | 203 | L/V |
| 1 | 7 | 508 | 67 | 63 | N/E |
| 1 | 7 | 430 | 48 | 44 | V/V |
| 1 | 7 | 2529 | 414 | 402 | R/R |
| 1 | 7 | 2493 | 406 | 394 | F/F |
| 1 | 7 | 1069 | 143 | 139 | T/T |
| 1 | 7 | 2581 | 420 | 408 | S/S |
| 1 | 7 | 2855 | 486 | 474 | K/K |
| 1 | 7 | 620 | 83 | 79 | F/F |
| 1 | 7 | 1398 | 197 | 193 | I/F |
| 2 | 14 | 1210 | 170 | 166 | S/E |
| 2 | 11 | 1208 | 168 | 164 | T/T |
| 2 | 10 | 2345 | 387 | 375 | V/V |
| 2 | 10 | 430 | 48 | 44 | V/V |
| 2 | 9 | 1024 | - | - | -/- |
| 2 | 9 | 550 | 70 | 66 | E/D |
| 2 | 9 | 2591 | 421 | 409 | S/S |
| 2 | 9 | 2391 | 392 | 380 | F/A |
| 2 | 9 | 1594 | 229 | 225 | E/E |
| 2 | 9 | 334 | 30 | 25 | F/F |
| 3 | 2 | 216 | 16 | 11 | D/D |
| 3 | 2 | 1380 | - | - | -/- |
| 3 | 2 | 865 | 110 | 106 | Q/N |
| 3 | 2 | 1607 | 232 | 228 | Q/Q |
| 3 | 2 | 2771 | - | - | -/- |
| 3 | 2 | 2600 | 424 | 412 | E/E |
| 3 | 2 | 601 | 77 | 73 | N/N |
| 3 | 2 | 993 | - | - | -/- |
| 3 | 2 | 1474 | 208 | 204 | F/I |
| 3 | 2 | 2827 | 479 | 467 | F/F |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mier, P.; Fontaine, J.-F.; Stoldt, M.; Libbrecht, R.; Martelli, C.; Foitzik, S.; Andrade-Navarro, M.A. Annotation and Analysis of 3902 Odorant Receptor Protein Sequences from 21 Insect Species Provide Insights into the Evolution of Odorant Receptor Gene Families in Solitary and Social Insects. Genes 2022, 13, 919. https://doi.org/10.3390/genes13050919
Mier P, Fontaine J-F, Stoldt M, Libbrecht R, Martelli C, Foitzik S, Andrade-Navarro MA. Annotation and Analysis of 3902 Odorant Receptor Protein Sequences from 21 Insect Species Provide Insights into the Evolution of Odorant Receptor Gene Families in Solitary and Social Insects. Genes. 2022; 13(5):919. https://doi.org/10.3390/genes13050919
Chicago/Turabian StyleMier, Pablo, Jean-Fred Fontaine, Marah Stoldt, Romain Libbrecht, Carlotta Martelli, Susanne Foitzik, and Miguel A. Andrade-Navarro. 2022. "Annotation and Analysis of 3902 Odorant Receptor Protein Sequences from 21 Insect Species Provide Insights into the Evolution of Odorant Receptor Gene Families in Solitary and Social Insects" Genes 13, no. 5: 919. https://doi.org/10.3390/genes13050919
APA StyleMier, P., Fontaine, J. -F., Stoldt, M., Libbrecht, R., Martelli, C., Foitzik, S., & Andrade-Navarro, M. A. (2022). Annotation and Analysis of 3902 Odorant Receptor Protein Sequences from 21 Insect Species Provide Insights into the Evolution of Odorant Receptor Gene Families in Solitary and Social Insects. Genes, 13(5), 919. https://doi.org/10.3390/genes13050919