TTC30A and TTC30B Redundancy Protects IFT Complex B Integrity and Its Pivotal Role in Ciliogenesis
Abstract
:1. Introduction
2. Materials and Methods
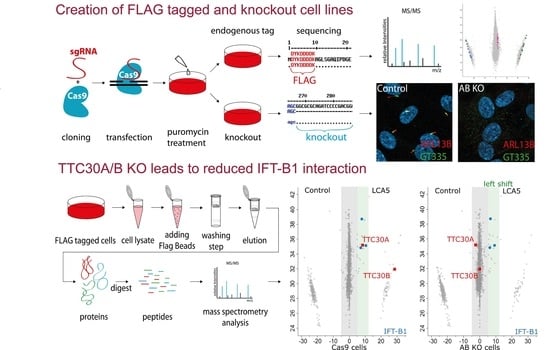
2.1. Generation of Knockin and Knockout Cell Lines
2.2. Immunofluorescence Staining
2.3. SDS-PAGE and WesternBlot
2.4. Affinity Purification
2.5. Mass Spectrometry
3. Results
3.1. Single TTC30A or TTC30B Knockout Mildly Affects Ciliogenesis with No Effect on IFT Complex A or B Component Localization
3.2. TTC30A and TTC30B Interact with IFT-B Complex Components
3.3. IFT-B Complex Composition Was Disturbed by Loss of TTC30A and TTC30B
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ishikawa, H.; Marshall, W.F. Ciliogenesis: Building the cell’s antenna. Nat. Rev. Mol. Cell. Biol. 2011, 12, 222–234. [Google Scholar] [CrossRef]
- Reiter, J.F.; Leroux, M.R. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 2017, 18, 533–547. [Google Scholar] [CrossRef] [PubMed]
- Wheway, G.; Schmidts, M.; Mans, D.A.; Szymanska, K.; Nguyen, T.M.T.; Racher, H.; Phelps, I.G.; Toedt, G.; Kennedy, J.; Wunderlich, K.A.; et al. An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat. Cell. Biol. 2015, 17, 1074–1087. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, J.L.; Witman, G.B. Intraflagellar transport. Nat. Rev. Mol. Cell. Biol. 2002, 3, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Scholey, J.M. Intraflagellar transport motors in cilia: Moving along the cell’s antenna. J. Cell Biol. 2008, 180, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Lechtreck, K.F. IFT–Cargo Interactions and Protein Transport in Cilia. Trends Biochem. Sci. 2015, 40, 765–778. [Google Scholar] [CrossRef] [Green Version]
- Cole, D.G.; Diener, D.R.; Himelblau, A.L.; Beech, P.L.; Fuster, J.C.; Rosenbaum, J.L. Chlamydomonas Kinesin-II–dependent Intraflagellar Transport (IFT): IFT Particles Contain Proteins Required for Ciliary Assembly in Caenorhabditis elegans Sensory Neurons. J. Cell Biol. 1998, 141, 993–1008. [Google Scholar] [CrossRef]
- Boldt, K.; van Reeuwijk, J.; Lu, Q.; Koutroumpas, K.; Nguyen, T.-M.T.; Texier, Y.; van Beersum, S.E.C.; Horn, N.; Willer, J.R.; Mans, D.A.; et al. An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat. Commun. 2016, 7, 11491. [Google Scholar] [CrossRef] [Green Version]
- Taschner, M.; Lorentzen, E. The Intraflagellar Transport Machinery. Cold Spring Harb. Perspect. Biol. 2016, 8, a028092. [Google Scholar] [CrossRef] [Green Version]
- Taschner, M.; Weber, K.; Mourão, A.; Vetter, M.; Awasthi, M.; Stiegler, M.; Bhogaraju, S.; Lorentzen, E. Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 2016, 35, 773–790. [Google Scholar] [CrossRef]
- Katoh, Y.; Terada, M.; Nishijima, Y.; Takei, R.; Nozaki, S.; Hamada, H.; Nakayama, K. Overall Architecture of the Intraflagellar Transport (IFT)-B Complex Containing Cluap1/IFT38 as an Essential Component of the IFT-B Peripheral Subcomplex. J. Biol. Chem. 2016, 291, 10962–10975. [Google Scholar] [CrossRef] [Green Version]
- Taschner, M.; Kotsis, F.; Braeuer, P.; Kuehn, E.W.; Lorentzen, E. Crystal structures of IFT70/52 and IFT52/46 provide insight into intraflagellar transport B core complex assembly. J. Cell Biol. 2014, 207, 269–282. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Chen, F.; Zhang, J.; Lin, Z.; Ma, Q.; Xu, G.; Xiao, D.; Gui, Y.; Yang, J.; Wan, S. A rare TTC30B variant is identified as a candidate for synpolydactyly in a Chinese pedigree. Bone 2019, 127, 503–509. [Google Scholar] [CrossRef]
- Takei, R.; Katoh, Y.; Nakayama, K. Robust interaction of IFT70 with IFT52–IFT88 in the IFT-B complex is required for ciliogenesis. Biol. Open 2018, 7, bio.033241. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.C.; Behal, R.H.; Geimer, S.; Wang, Z.; Williamson, S.M.; Zhang, H.; Cole, D.G.; Qin, H. Chlamydomonas IFT70/CrDYF-1 is a core component of IFT particle complex B and is required for flagellar assembly. Mol. Biol. Cell 2010, 21, 2696–2706. [Google Scholar] [CrossRef] [Green Version]
- Taschner, M.; Bhogaraju, S.; Vetter, M.; Morawetz, M.; Lorentzen, E. Biochemical mapping of interactions within the intraflagellar transport (IFT) B core complex: IFT52 binds directly to four other IFT-B subunits. J. Biol. Chem. 2011, 286, 26344–26352. [Google Scholar] [CrossRef] [Green Version]
- Pathak, N.; Obara, T.; Mangos, S.; Liu, Y.; Drummond, I.A. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol. Biol. Cell 2007, 18, 4353–4364. [Google Scholar] [CrossRef] [Green Version]
- Bobinnec, Y.; Khodjakov, A.; Mir, L.; Rieder, C.; Eddé, B.; Bornens, M. Centriole Disassembly In Vivo and Its Effect on Centrosome Structure and Function in Vertebrate Cells. J. Cell Biol. 1998, 143, 1575–1589. [Google Scholar] [CrossRef] [Green Version]
- Redeker, V.; Levilliers, N.; Vinolo, E.; Rossier, J.; Jaillard, D.; Burnette, D.; Gaertig, J.; Bré, M.H. Mutations of tubulin glycylation sites reveal cross-talk between the C termini of alpha- and beta-tubulin and affect the ciliary matrix in Tetrahymena. J. Biol. Chem. 2005, 280, 596–606. [Google Scholar] [CrossRef] [Green Version]
- Pathak, N.; Austin, C.A.; Drummond, I.A. Tubulin Tyrosine Ligase-like Genes ttll3 and ttll6 Maintain Zebrafish Cilia Structure and Motility. J. Biol. Chem. 2011, 286, 11685–11695. [Google Scholar] [CrossRef] [Green Version]
- Pathak, N.; Austin-Tse, C.; Liu, Y.; Vasilyev, A.; Drummond, I.A. Cytoplasmic carboxypeptidase 5 regulates tubulin glutamylation and zebrafish cilia formation and function. Mol. Biol. Cell 2014, 25, 1836–1844. [Google Scholar] [CrossRef]
- Howard, P.W.; Jue, S.F.; Maurer, R.A. Interaction of mouse TTC30/DYF-1 with multiple intraflagellar transport complex B proteins and KIF17. Exp. Cell Res. 2013, 319, 2275–2281. [Google Scholar] [CrossRef] [Green Version]
- Stemmer, M.; Thumberger, T.; Keyer, M.D.S.; Wittbrodt, J.; Mateo, J.L. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS ONE 2015, 10, e0124633. [Google Scholar] [CrossRef] [Green Version]
- Labuhn, M.; Adams, F.F.; Ng, M.; Knoess, S.; Schambach, A.; Charpentier, E.M.; Schwarzer, A.; Mateo, J.L.; Klusmann, J.-H.; Heckl, D. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR–Cas9 applications. Nucleic Acids Res. 2018, 46, 1375–1385. [Google Scholar] [CrossRef] [Green Version]
- Meijering, E.; Jacob, M.; Sarria, J.-C.F.; Steiner, P.; Hirling, H.; Unser, M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry 2004, 58A, 167–176. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Dummer, A.; Poelma, C.; DeRuiter, M.C.; Goumans, M.-J.T.H.; Hierck, B.P. Measuring the primary cilium length: Improved method for unbiased high-throughput analysis. Cilia 2016, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Ling, K.; Hu, J. The emerging role of tubulin posttranslational modifications in cilia and ciliopathies. Biophys. Rep. 2020, 6, 89–104. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Boldt, K.; Mans, R.A.; Won, J.; Van Reeuwijk, J.; Vogt, A.; Kinkl, N.; Letteboer, S.J.; Hicks, W.L.; Hurd, R.E.; Naggert, J.K.; et al. Disruption of intraflagellar protein transport in photoreceptor cilia causes Leber congenital amaurosis in humans and mice. J. Clin. Investig. 2011, 121, 2169–2180. [Google Scholar] [CrossRef] [PubMed]
- Beyer, T.; Klose, F.; Kuret, A.; Hoffmann, F.; Lukowski, R.; Ueffing, M.; Boldt, K. Tissue- and isoform-specific protein complex analysis with natively processed bait proteins. J. Proteom. 2021, 231, 103947. [Google Scholar] [CrossRef] [PubMed]
- Gloeckner, C.J.; Boldt, K.; Ueffing, M. Strep/FLAG Tandem Affinity Purification (SF-TAP) to Study Protein Interactions. Curr. Protoc. Protein Sci. 2009, 57, 19–20. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kim, H.; Lee, S. Mismatch Intolerance of 5′-Truncated sgRNAs in CRISPR/Cas9 Enables Efficient Microbial Single-Base Genome Editing. Int. J. Mol. Sci. 2021, 22, 6457. [Google Scholar] [CrossRef]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2019, 9, 1911. [Google Scholar] [CrossRef]
- Adli, M. The Biology and Application Areas of CRISPR Technologies. J. Mol. Biol. 2019, 431, 1–2. [Google Scholar] [CrossRef]
- Simon, R.; Durrleman, S.; Hoppe, R.T.; Bonadonna, G.; Bloomfield, C.D.; Rudders, R.A.; Cheson, B.D.; Berard, C.W. The Non-Hodgkin Lymphoma Pathologic Classification Project. Long-term follow-up of 1153 patients with non-Hodgkin lymphomas. Ann. Intern. Med. 1988, 109, 939–945. [Google Scholar] [CrossRef]
- Gibson, T.J.; Seiler, M.; A Veitia, R. The transience of transient overexpression. Nat. Methods 2013, 10, 715–721. [Google Scholar] [CrossRef]
- Xu, M.; Xie, Y.A.; Abouzeid, H.; Gordon, C.T.; Fiorentino, A.; Sun, Z.; Lehman, A.; Osman, I.S.; Dharmat, R.; Riveiro-Alvarez, R.; et al. Mutations in the Spliceosome Component CWC27 Cause Retinal Degeneration with or without Additional Developmental Anomalies. Am. J. Hum. Genet. 2017, 100, 592–604. [Google Scholar] [CrossRef] [Green Version]
- Bahe, S.; Stierhof, Y.-D.; Wilkinson, C.; Leiss, F.; Nigg, E.A. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 2005, 171, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Gao, J.; Adamian, M.; Wen, X.-H.; Pawlyk, B.; Zhang, L.; Sanderson, M.J.; Zuo, J.; Makino, C.L.; Li, T. The Ciliary Rootlet Maintains Long-Term Stability of Sensory Cilia. Mol. Cell. Biol. 2005, 25, 4129–4137. [Google Scholar] [CrossRef] [Green Version]
- Mohan, S.; Timbers, T.A.; Kennedy, J.; Blacque, O.E.; Leroux, M.R. Striated Rootlet and Nonfilamentous Forms of Rootletin Maintain Ciliary Function. Curr. Biol. 2013, 23, 2016–2022. [Google Scholar] [CrossRef] [Green Version]
- Jühlen, R.; Martinelli, V.; Vinci, C.; Breckpot, J.; Fahrenkrog, B. Centrosome and ciliary abnormalities in fetal akinesia deformation sequence human fibroblasts. Sci. Rep. 2020, 10, 19301. [Google Scholar] [CrossRef]
- Watanabe, Y.; Nakagawa, T.; Akiyama, T.; Nakagawa, M.; Suzuki, N.; Warita, H.; Aoki, M.; Nakayama, K. An Amyotrophic Lateral Sclerosis-Associated Mutant of C21ORF2 Is Stabilized by NEK1-Mediated Hyperphosphorylation and the Inability to Bind FBXO3. iScience 2020, 23, 101491. [Google Scholar] [CrossRef]
- Berbari, N.F.; Kin, N.W.; Sharma, N.; Michaud, E.J.; Kesterson, R.A.; Yoder, B.K. Mutations in Traf3ip1 reveal defects in ciliogenesis, embryonic development, and altered cell size regulation. Dev. Biol. 2011, 360, 66–76. [Google Scholar] [CrossRef] [Green Version]
- Pasek, R.C.; Berbari, N.F.; Lewis, W.R.; Kesterson, R.A.; Yoder, B.K. Mammalian Clusterin associated protein 1 is an evolutionarily conserved protein required for ciliogenesis. Cilia 2012, 1, 20. [Google Scholar] [CrossRef] [Green Version]
- Quélin, C.; Loget, P.; Boutaud, L.; Elkhartoufi, N.; Milon, J.; Odent, S.; Fradin, M.; Demurger, F.; Pasquier, L.; Thomas, S.; et al. Loss of function IFT27 variants associated with an unclassified lethal fetal ciliopathy with renal agenesis. Am. J. Med Genet. Part A 2018, 176, 1610–1613. [Google Scholar] [CrossRef]
- Getwan, M.; Hoppmann, A.; Schlosser, P.; Grand, K.; Song, W.; Diehl, R.; Schroda, S.; Heeg, F.; Deutsch, K.; Hildebrandt, F.; et al. Ttc30a affects tubulin modifications in a model for ciliary chondrodysplasia with polycystic kidney disease. Proc. Natl. Acad. Sci. USA 2021, 118, e2106770118. [Google Scholar] [CrossRef]
- He, K.; Ma, X.; Xu, T.; Li, Y.; Hodge, A.; Zhang, Q.; Torline, J.; Huang, Y.; Zhao, J.; Ling, K.; et al. Axoneme polyglutamylation regulated by Joubert syndrome protein ARL13B controls ciliary targeting of signaling molecules. Nat. Commun. 2018, 9, 3310. [Google Scholar] [CrossRef] [Green Version]
- Kanamaru, T.; Neuner, A.; Kurtulmus, B.; Pereira, G. Balancing the length of the distal tip by septins is key for stability and signalling function of primary cilia. EMBO J. 2022, 41, e108843. [Google Scholar] [CrossRef]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]







| Bait | LCA5 | RAF1 | LCA5 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Cell Line | TTC30A/B KO versus Control | TTC30B KO versus Control | TTC30 A/B KO | TTC30 B KO | Cas Control | TTC30 A/B KO | TTC30 B KO | Cas Control | |
| Gene Name | |||||||||
| TTC30B | −28.42 | −28.42 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 28.42 | |
| TTC30A | −10.88 | 0.42 | 0.00 | 0.00 | 22.36 | 19.87 | 31.17 | 30.75 | |
| IFT52 | −1.13 | 0.32 | 0.00 | 0.00 | 20.46 | 29.69 | 31.14 | 30.82 | |
| IFT46 | −1.04 | 0.45 | 21.56 | 21.85 | 23.18 | 29.37 | 30.87 | 30.42 | |
| IFT81 | −1.44 | 0.14 | 20.64 | 23.03 | 22.58 | 30.79 | 32.37 | 32.24 | |
| IFT74 | −1.32 | 0.14 | 24.04 | 24.01 | 24.30 | 30.86 | 32.32 | 32.18 | |
| IFT88 | −1.27 | 0.57 | 19.93 | 0.00 | 23.55 | 30.16 | 32.00 | 31.43 | |
| TTC26 | −1.24 | 0.21 | 23.99 | 23.94 | 24.05 | 29.66 | 31.12 | 30.91 | |
| IFT27 | −1.03 | 0.17 | 25.32 | 25.19 | 25.27 | 29.51 | 30.71 | 30.54 | |
| IFT22 | −1.02 | 0.26 | 0.00 | 23.17 | 22.89 | 29.69 | 30.97 | 30.71 | |
| HSPB11 | −0.94 | 0.26 | 24.00 | 23.97 | 24.24 | 28.20 | 29.40 | 29.13 | |
| TRAF3IP1 | −0.34 | 0.88 | 23.45 | 22.69 | 21.40 | 29.91 | 31.13 | 30.24 | |
| CLUAP1 | −0.33 | 0.43 | 0.00 | 0.00 | 18.33 | 31.27 | 32.03 | 31.60 | |
| IFT57 | −0.27 | 0.46 | 21.63 | 21.16 | 20.58 | 31.54 | 32.27 | 31.81 | |
| IFT80 | −0.20 | 0.53 | 18.97 | 0.00 | 0.00 | 31.96 | 32.70 | 32.16 | |
| IFT172 | −0.18 | 0.46 | 24.82 | 24.41 | 24.62 | 34.01 | 34.64 | 34.18 | |
| IFT20 | −0.06 | 0.60 | 0.00 | 0.00 | 0.00 | 30.93 | 31.60 | 31.00 | |
| LCA5 | 0.47 | 0.42 | 24.86 | 24.98 | 24.46 | 37.55 | 37.50 | 37.08 | |
| Bait | IFT88 | RAF1 | IFT88 | ||||||
| Cell Line | TTC30A/B KO versus Control | TTC30B KO versus Control | TTC30 A/B KO | TTC30 B KO | Cas Control | TTC30 A/B KO | TTC30 B KO | Cas Control | |
| Gene Name | |||||||||
| TTC30B | −26.77 | −26.77 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 26.77 | |
| TTC30A | −7.47 | −0.67 | 0.00 | 0.00 | 22.36 | 21.64 | 28.44 | 29.11 | |
| IFT52 | −1.22 | −0.60 | 0.00 | 0.00 | 20.46 | 27.58 | 28.20 | 28.80 | |
| IFT46 | −1.19 | −0.59 | 21.56 | 21.85 | 23.18 | 27.44 | 28.04 | 28.63 | |
| IFT81 | −1.34 | −0.67 | 20.64 | 23.03 | 22.58 | 28.77 | 29.45 | 30.11 | |
| IFT74 | −1.49 | −0.80 | 24.04 | 24.01 | 24.30 | 28.69 | 29.38 | 30.18 | |
| IFT88 | −0.40 | −0.46 | 19.93 | 0.00 | 23.55 | 34.18 | 34.11 | 34.57 | |
| TTC26 | −1.09 | −0.59 | 23.99 | 23.94 | 24.05 | 27.86 | 28.36 | 28.94 | |
| IFT27 | −0.98 | −0.67 | 25.32 | 25.19 | 25.27 | 27.68 | 28.00 | 28.67 | |
| IFT22 | −1.29 | −0.81 | 0.00 | 23.17 | 22.89 | 27.55 | 28.03 | 28.85 | |
| HSPB11 | −0.94 | −0.61 | 24.00 | 23.97 | 24.24 | 26.43 | 26.76 | 27.37 | |
| TRAF3IP1 | 0.78 | −0.03 | 23.45 | 22.69 | 21.40 | 25.57 | 24.75 | 24.79 | |
| CLUAP1 | 0.34 | −0.21 | 0.00 | 0.00 | 18.33 | 26.22 | 25.67 | 25.88 | |
| IFT57 | 0.46 | −0.24 | 21.63 | 21.16 | 20.58 | 26.74 | 26.03 | 26.28 | |
| IFT80 | 0.77 | 0.03 | 18.97 | 0.00 | 0.00 | 26.05 | 25.30 | 25.28 | |
| IFT172 | 0.41 | −0.10 | 24.82 | 24.41 | 24.62 | 29.33 | 28.82 | 28.92 | |
| IFT20 | 0.56 | −0.10 | 0.00 | 0.00 | 0.00 | 26.22 | 25.56 | 25.66 | |
| LCA5 | 0.75 | −0.27 | 24.86 | 24.98 | 24.46 | 26.62 | 25.60 | 25.88 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoffmann, F.; Bolz, S.; Junger, K.; Klose, F.; Schubert, T.; Woerz, F.; Boldt, K.; Ueffing, M.; Beyer, T. TTC30A and TTC30B Redundancy Protects IFT Complex B Integrity and Its Pivotal Role in Ciliogenesis. Genes 2022, 13, 1191. https://doi.org/10.3390/genes13071191
Hoffmann F, Bolz S, Junger K, Klose F, Schubert T, Woerz F, Boldt K, Ueffing M, Beyer T. TTC30A and TTC30B Redundancy Protects IFT Complex B Integrity and Its Pivotal Role in Ciliogenesis. Genes. 2022; 13(7):1191. https://doi.org/10.3390/genes13071191
Chicago/Turabian StyleHoffmann, Felix, Sylvia Bolz, Katrin Junger, Franziska Klose, Timm Schubert, Franziska Woerz, Karsten Boldt, Marius Ueffing, and Tina Beyer. 2022. "TTC30A and TTC30B Redundancy Protects IFT Complex B Integrity and Its Pivotal Role in Ciliogenesis" Genes 13, no. 7: 1191. https://doi.org/10.3390/genes13071191
APA StyleHoffmann, F., Bolz, S., Junger, K., Klose, F., Schubert, T., Woerz, F., Boldt, K., Ueffing, M., & Beyer, T. (2022). TTC30A and TTC30B Redundancy Protects IFT Complex B Integrity and Its Pivotal Role in Ciliogenesis. Genes, 13(7), 1191. https://doi.org/10.3390/genes13071191