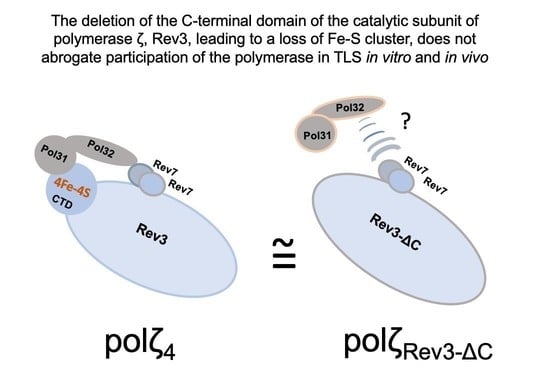
DNA Polymerase ζ without the C-Terminus of Catalytic Subunit Rev3 Retains Characteristic Activity, but Alters Mutation Specificity of Ultraviolet Radiation in Yeast
Abstract
:1. Introduction
2. Materials and Methods
2.1. Nomenclature and Abbreviations
2.2. Yeast Strains, Media, and Plasmids
2.3. Polymerase Purification
2.4. DNA Substrates
2.5. Determination of Survival and Induced Mutation Frequencies Using Canr Assay
2.6. Analysis of Mutational Spectra
2.7. Statistical Analyses
3. Results
3.1. Pol ζRev3ΔC Shows Hallmark Characteristics of the TLS Polymerase pol ζ4, but Has Lower Activity In Vitro
3.2. Rev3-∆C ιs Most Proficient at Participating in the Formation of Transition Mutations In Vivo
3.2.1. Dose of 20 J/m2
3.2.2. Dose of 40 J/m2
3.2.3. Dose of 60 J/m2
3.3. Pol ζRev3ΔC Participates in Creating pol ζ-specific Complex Mutations with Reduced Efficiency
3.4. The Majority of Mutations in Wild-type and rev3-∆C Strains Occur in Dipyrimidine (DP) Sites
4. Discussion
4.1. Biochemical Properties of pol ζRev3ΔC.
4.2. UVR Mutation Spectra in rev3-∆C Strains
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Friedberg, E.C. Suffering in silence: The tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 2005, 6, 943–953. [Google Scholar] [CrossRef]
- Sinha, R.P.; Hader, D. UV-induced DNA damage and repair: A review. R. Soc. Chem. Own. Soc. 2002, 1, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Yang, W. Surviving the Sun: Repair and bypass of DNA UV lesions. Protein Sci. 2011, 20, 1781. [Google Scholar] [CrossRef] [PubMed]
- Cleaver, J.E. Mechanisms by which human cells bypass damaged bases during DNA replication after ultraviolet irradiation. Sci. World J. 2002, 2, 1296–1305. [Google Scholar] [CrossRef] [PubMed]
- de Gruijl, F.R. Skin cancer and solar UV radiation. Eur. J. Cancer 1999, 35, 2003–2009. [Google Scholar] [CrossRef]
- Narayanan, D.L.; Saladi, R.N.; Fox, J.L. Ultraviolet radiation and skin cancer. Int. J. Dermatol. 2010, 49, 978–986. [Google Scholar] [CrossRef]
- Ravanat, J.; Douki, T.; Cadet, J. Direct and indirect effects of UV radiation on DNA and its components. J. Photochem. Photobiol. B Biol. 2001, 63, 88–102. [Google Scholar] [CrossRef]
- Solomon, K.R. Effects of ozone depletion and UV-B radiation on humans and the environment. Atmos.-Ocean. 2008, 46, 185–202. [Google Scholar] [CrossRef]
- Kerr, J.B.; McElroy, C.T. Evidence for large upward trends of ultraviolet-B radiation linked to ozone depletion. Science 1993, 262, 1032–1034. [Google Scholar] [CrossRef]
- Mullenders, L.H.F. Solar UV damage to cellular DNA: From mechanisms to biological effects. Photochem. Photobiol. Sci. 2018, 17, 1842–1852. [Google Scholar] [CrossRef]
- Friedberg, E.C.; Walker, G.C.; Siede, W.; Wood, R.D.; Schultz, R.A.; Ellenberger, T. DNA Repair and Mutagenesis, 2nd ed.; ASM Press: Washington, DC, USA, 2006. [Google Scholar]
- Brash, D.E.; Haseltine, W.A. UV-induced mutation hotspots occur at DNA damage hotspots. Nature 1982, 298, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Ait Saada, A.; Lambert, S.A.E.; Carr, A.M. Preserving replication fork integrity and competence via the homologous recombination pathway. DNA Repair. 2018, 71, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.; Johnson, R.E.; Prakash, L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem. 2005, 74, 317–353. [Google Scholar] [CrossRef] [PubMed]
- Vaisman, A.; Woodgate, R. Translesion DNA polymerases in eukaryotes: What makes them tick? Crit. Rev. Biochem. Mol. Biol. 2017, 52, 274–303. [Google Scholar] [CrossRef]
- Yang, W.; Gao, Y. Translesion and Repair DNA Polymerases: Diverse structure and mechanism. Annu. Rev. Biochem. 2018, 87, 239–261. [Google Scholar] [CrossRef]
- Johnson, R.E.; Prakash, S.; Prakash, L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, polη. Science 1999, 283, 1001–1004. [Google Scholar] [CrossRef]
- McCulloch, S.D.; Kokoska, R.J.; Masutani, C.; Iwai, S.; Hanaoka, F.; Kunkel, T.A. Preferential cis-syn thymine dimer bypass by DNA polymerase η occurs with biased fidelity. Nature 2004, 428, 87–100. [Google Scholar] [CrossRef]
- Abdulovic, A.L.; Jinks-Robertson, S. The in vivo characterization of translesion synthesis across UV-induced lesions in Saccharomyces cerevisiae: Insights into polζ- and polη-dependent frameshift mutagenesis. Genetics 2006, 172, 1487–1498. [Google Scholar] [CrossRef]
- Kozmin, S.G.; Pavlov, Y.I.; Kunkel, T.A.; Sage, E. Roles of Saccharomyces cerevisiae DNA polymerases polη and polζ in response to irradiation by simulated sunlight. Nucleic Acids Res. 2003, 31, 4541–4552. [Google Scholar] [CrossRef]
- Gibbs, P.E.; McDonald, J.; Woodgate, R.; Lawrence, C.W. The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 2005, 169, 575–582. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, C.W.; Maher, V.M. Mutagenesis in eukaryotes dependent on DNA polymerase ζ and Rev1p. Philos. Trans. R. Soc. Lond. 2001, 356, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Makarova, A.V.; Burgers, P.M. Eukaryotic DNA polymerase ζ. DNA Repair 2015, 29, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Waters, L.S.; Minesinger, B.K.; Wiltrout, M.E.; D’Souza, S.; Woodruff, R.V.; Walker, G.C. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. MMBR 2009, 73, 134–154. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Garg, P.; Stith, C.M.; Nick McElhinny, S.A.; Kissling, G.E.; Burgers, P.M.; Kunkel, T.A. The fidelity of DNA synthesis by yeast DNA polymerase ζ alone and with accessory proteins. Nucleic Acids Res. 2006, 34, 4731–4742. [Google Scholar] [CrossRef]
- Kochenova, O.V.; Bezalel-Buch, R.; Tran, P.; Makarova, A.V.; Chabes, A.; Burgers, P.M.; Shcherbakova, P.V. Yeast DNA polymerase zeta maintains consistent activity and mutagenicity across a wide range of physiological dNTP concentrations. Nucleic Acids Res. 2017, 45, 1200–1218. [Google Scholar] [CrossRef]
- Northam, M.R.; Garg, P.; Baitin, D.M.; Burgers, P.M.; Shcherbakova, P.V. A Novel Function of DNA Polymerase ζ Regulated by PCNA. EMBO J. 2006, 25, 4316–4325. [Google Scholar] [CrossRef]
- Abdulovic, A.L.; Minesinger, B.K.; Jinks-Robertson, S. The effect of sequence context on spontaneous Pol zeta-dependent mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 2008, 36, 2082–2093. [Google Scholar] [CrossRef]
- Northam, M.R.; Robinson, H.A.; Kochenova, O.V.; Shcherbakova, P.V. Participation of DNA polymerase ζ in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics 2010, 184, 27–42. [Google Scholar] [CrossRef]
- Garbacz, M.; Araki, H.; Flis, K.; Bebenek, A.; Zawada, A.E.; Jonczyk, P.; Makiela-Dzbenska, K.; Fijalkowska, I.J. Fidelity consequences of the impaired interaction between DNA polymerase epsilon and the GINS complex. DNA Repair 2015, 29, 23–35. [Google Scholar] [CrossRef]
- Szwajczak, E.; Fijalkowska, I.J.; Suski, C. The CysB motif of Rev3p involved in the formation of the four-subunit DNA polymerase ζ is required for defective-replisome-induced mutagenesis. Mol. Microbiol. 2017, 106, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Kochenova, O.V.; Daee, D.L.; Mertz, T.M.; Shcherbakova, P.V. DNA polymerase ζ-dependent lesion bypass in Saccharomyces cerevisiae is accompanied by error-prone copying of long stretches of adjacent DNA. PLoS Genet. 2015, 11, e1005110. [Google Scholar] [CrossRef] [PubMed]
- Northam, M.R.; Moore, E.A.; Mertz, T.M.; Binz, S.K.; Stith, C.M.; Stepchenkova, E.I.; Wendt, K.L.; Burgers, P.M.; Shcherbakova, P.V. DNA polymerases ζ and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Res. 2014, 42, 290–306. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.R.; Lawrence, C.W.; Hinkle, D. Thymine-Thymine Dimer Bypass by Yeast DNA Polymerase ζ. Science 1996, 272, 1646–1649. [Google Scholar] [CrossRef]
- Prakash, S.; Prakash, L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genome Res. 2002, 16, 1872–1883. [Google Scholar] [CrossRef]
- Pages, V.; Johnson, R.E.; Prakash, L.; Prakash, S. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc. Natl. Acad. Sci. USA 2008, 105, 1170–1175. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.P.; Levine, A.S.; Woodgate, R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 1997, 147, 1557–1568. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, Y.I.; Zhuk, A.S.; Stepchenkova, E.I. DNA Polymerases at the Eukaryotic Replication Fork Thirty Years after: Connection to Cancer. Cancers 2020, 12, 3489. [Google Scholar] [CrossRef]
- Baranovskiy, A.G.; Lada, A.G.; Siebler, H.M.; Zhang, Y.; Pavlov, Y.I.; Tahirov, T.H. DNA Polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase Δ. J. Biol. Chem. 2012, 287, 17281–17287. [Google Scholar] [CrossRef]
- Johnson, R.E.; Prakash, L.; Prakash, S. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc. Natl. Acad. Sci. USA 2012, 109, 12455–12460. [Google Scholar] [CrossRef]
- Makarova, A.V.; Stodola, J.L.; Burgers, P.M. A four-subunit DNA polymerase zeta complex containing pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012, 40, 11618–11626. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.S.; Gregory, M.T.; Yang, W. Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc. Natl. Acad. Sci. USA 2014, 111, 2954–2959. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.K.; Wood, R.D. DNA polymerase ζ in DNA replication and repair. Nucleic Acids Res. 2019, 47, 8348–8361. [Google Scholar] [CrossRef] [PubMed]
- Malik, R.; Kopylov, M.; Gomez-Llorente, Y.; Jain, R.; Johnson, R.E.; Prakash, L.; Prakash, S.; Ubarretxena-Belandia, I.; Aggarwal, A.K. Structure and mechanism of B-family DNA polymerase ζ specialized for translesion DNA synthesis. Nat. Struct. Mol. Biol. 2020, 27, 913–924. [Google Scholar] [CrossRef]
- Waisertreiger, I.S.; Liston, V.G.; Menezes, M.R.; Kim, H.M.; Lobachev, K.S.; Stepchenkova, E.I.; Tahirov, T.H.; Rogozin, I.B.; Pavlov, Y.I. Modulation of mutagenesis in eukaryotes by DNA replication fork dynamics and quality of nucleotide pools. Environ. Mol. Mutagenesis 2012, 53, 699–724. [Google Scholar] [CrossRef] [PubMed]
- Baranovskiy, A.G.; Babayeva, N.D.; Liston, V.G.; Rogozin, I.B.; Koonin, E.V.; Pavlov, Y.I.; Vassylyev, D.G.; Tahirov, T.H. X-ray structure of the complex of regulatory subunits of human DNA polymerase delta. Cell Cycle 2008, 7, 3026–3036. [Google Scholar] [CrossRef]
- Siebler, H.M.; Lada, A.G.; Baranovskiy, A.G.; Tahirov, T.H.; Pavlov, Y.I. A novel variant of DNA polymerase ζ, Rev3ΔC, highlights differential regulation of Pol32 as a subunit of polymerase δ versus ζ in Saccharomyces cerevisiae. DNA Repair 2014, 24, 138–149. [Google Scholar] [CrossRef]
- Fortune, J.M.; Pavlov, Y.I.; Welch, C.M.; Johansson, E.; Burgers, P.M.; Kunkel, T.A. Saccharomyces cerevisiae DNA polymerase δ: High fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005, 280, 29980–29987. [Google Scholar] [CrossRef]
- Pavlov, Y.I.; Shcherbakova, P.V.; Kunkel, T.A. In vivo consequences of putative active site mutations in yeast DNA polymerases α, ε, δ, and ζ. Genetics 2001, 159, 47–64. [Google Scholar] [CrossRef]
- Stepchenkova, E.I.; Zhuk, A.S.; Cui, J.; Tarakhovskaya, E.R.; Barbari, S.R.; Shcherbakova, P.V.; Polev, D.E.; Fedorov, R.; Poliakov, E.; Rogozin, I.B.; et al. Compensation for the absence of the catalytically active half of DNA polymerase ε in yeast by positively selected mutations in CDC28. Genetics 2021, 218, lyab060. [Google Scholar] [CrossRef]
- Zhang, Y.; Baranovskiy, A.G.; Tahirov, E.T.; Tahirov, T.H.; Pavlov, Y.I. Divalent ions attenuate DNA synthesis by human DNA polymerase α by changing the structure of the template/primer or by perturbing the polymerase reaction. DNA Repair 2016, 43, 24–33. [Google Scholar] [CrossRef] [Green Version]
- Kozmin, S.G.; Rogozin, I.B.; Moore, E.A.; Abney, M.; Schaaper, R.M.; Pavlov, Y.I. Comment on “A commensal strain of Staphylococcus epidermidis protects against skin neoplasia” by Nakatsuji et al. Sci. Adv. 2019, 5, eaaw3915. [Google Scholar] [CrossRef] [PubMed]
- Lis, J.T.; Schleif, R. Size fractionation of double-stranded DNA by precipitation with polyethylene glycol. Nucleic Acids Res. 1975, 2, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Wojtaszek, J.L.; Chatterjee, N.; Najeeb, J.; Ramos, A.; Lee, M.; Bian, K.; Xue, J.Y.; Fenton, B.A.; Park, H.; Li, D.; et al. A small molecule targeting mutagenic translesion synthesis improves chemotherapy. Cell 2019, 178, 152–159.e111. [Google Scholar] [CrossRef]
- Malik, R.; Johnson, R.E.; Prakash, L.; Prakash, S.; Ubarretxena-Belandia, I.; Aggarwal, A.K. Cryo-EM structure of translesion DNA synthesis polymerase ζ with a base pair mismatch. Nat. Commun. 2022, 13, 1050. [Google Scholar] [CrossRef] [PubMed]
- Acharya, N.; Johnson, R.E.; Pagès, V.; Prakash, L.; Prakash, S. Yeast Rev1 protein promotes complex formation of DNA polymerase zeta with Pol32 subunit of DNA polymerase delta. Proc. Natl. Acad. Sci. USA 2009, 106, 9631–9636. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, S.; Hara, K.; Shimizu, T.; Sato, M.; Hashimoto, H. Structural basis of recruitment of DNA polymerase ζ by interaction between REV1 and REV7 proteins. J. Biol. Chem. 2012, 287, 33847–33852. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, A.A.; Korzhnev, D.M. The Rev1-Polζ translesion synthesis mutasome: Structure, interactions and inhibition. Enzymes 2019, 45, 139–181. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Malyarchuk, B.A.; Pavlov, Y.I.; Milanesi, L. From context-dependence of mutations to molecular mechanisms of mutagenesis. In Biocomputing 2005, Proceedings of the Pacific Symposium, Hawaii, HI, USA, 4–8 January 2005; World Secentific: Singapore, 2005; pp. 409–420. [Google Scholar]
- Dogliotti, E.; Hainaut, P.; Hernandez, T.; D’Errico, M.; DeMarini, D.M. Mutation spectra resulting from carcinogenic exposure: From model systems to cancer-related genes. Recent Results Cancer Res. 1998, 154, 97–124. [Google Scholar]
- Giglia-Mari, G.; Sarasin, A. TP53 Mutations in Human Skin Cancers. Hum. Mutat. 2003, 21, 217–228. [Google Scholar] [CrossRef]
- Daya-Grosjean, L.; Dumaz, N.; Sarasin, A. The specificity of p53 mutation spectra in sunlight induced human cancers. J. Photochem. Photobiol. 1995, 28, 115–124. [Google Scholar] [CrossRef]
- Inga, A.; Scott, G.; Monti, P.; Aprile, A.; Abbondandolo, A.; Burns, P.A.; Fronza, G. Ultraviolet-light induced p53 mutational spectrum in yeast is indistinguishable from p53 mutations in human skin cancer. Carcinogenesis 1998, 19, 741–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harfe, B.D.; Jinks-Robertson, S. DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell 2000, 6, 1491–1499. [Google Scholar] [CrossRef]
- Boiteux, S.; Jinks-Robertson, S. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics 2013, 193, 1025–1064. [Google Scholar] [CrossRef] [PubMed]
- Burger, A.; Fix, D.; Liu, H.; Hays, J.; Bockrath, R. In vivo deamination of cytosine-containing cyclobutane pyrimidine dimers in E. coli: A feasible part of UV-mutagenesis. Mutat. Res. 2003, 522, 145–156. [Google Scholar] [CrossRef]
- Holt, M.E.; Salay, L.E.; Chazin, W.J. A polymerase with potential: The Fe-S cluster in human DNA primase. Methods Enzymol. 2017, 595, 361–390. [Google Scholar] [CrossRef]
- Bartels, P.L.; Stodola, J.L.; Burgers, P.M.J.; Barton, J.K. A Redox Role for the [4Fe4S] Cluster of yeast DNA polymerase δ. J. Am. Chem. Soc. 2017, 139, 18339–18348. [Google Scholar] [CrossRef]
- Pinto, M.N.; Ter Beek, J.; Ekanger, L.A.; Johansson, E.; Barton, J.K. The [4Fe4S] cluster of yeast DNA polymerase ε is redox active and can undergo DNA-mediated signaling. J. Am. Chem. Soc. 2021, 143, 16147–16153. [Google Scholar] [CrossRef]





| Mutation Type * | UVR dose (J/m2) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 20 | 40 | 60 | |||||||
| Wild-Type | rev3-ΔC | p-Value # | Wild-Type | rev3-ΔC | p-Value # | Wild-Type | rev3-ΔC | p-Value # | |
| transitions | |||||||||
| AT to GC | 6 | 6 | 6 | 10 | 8 | 1# | 0.0386 | ||
| GC to AT | 17 | 20 | 13 | 23# | 21 | 41# | <0.0001 | ||
| Total | 23 | 26 | 19 | 33# | 29 | 42# | 0.0007 | ||
| Rate ** | 3.5 | 6.8 | 9.9 | 9.1 | 29.0 | 4.5 | |||
| Fold decrease ## | 0.5 | 1.1 | 6.4 | ||||||
| Transversions | |||||||||
| AT to TA | 18 | 14 | 19 | 8# | 0.0206 | 15 | 6 | ||
| Other | 2 | 0 | 7 | 6 | 8 | 4 | |||
| Total | 20 | 14 | 26 | 14# | 0.0132 | 23 | 10# | 0.0445 | |
| Rate ** | 3.0 | 3.7 | 13.6 | 3.9 | 24.0 | 1.1 | |||
| Fold decrease ## | 0.8 | 3.5 | 22.3 | ||||||
| Frameshifts | |||||||||
| Deletions | 10 | 8 | 10 | 9 | 16 | 5# | 0.0322 | ||
| Insertions | 3 | 1 | 2 | 3 | 1 | 1 | |||
| Total | 13 | 9 | 12 | 12 | 17 | 6# | 0.0407 | ||
| Rate ** | 2.0 | 2.4 | 6.2 | 3.3 | 16.0 | 0.6 | |||
| Fold decrease ## | 0.8 | 1.9 | 24.8 | ||||||
| Total changes | 56 | 49 | 57 | 59 | 69 | 58 | |||
| Total rate ** | 8.5 | 12.8 | 29.7 | 16.4 | 69.0 | 6.25 | |||
| Fold decrease ## | 0.7 | 1.8 | 11.1 | ||||||
| DP Site | UVR Dose, J/m2 | |||||
|---|---|---|---|---|---|---|
| 20 | 40 | 60 | ||||
| Wild-Type | rev3-ΔC | Wild-Type | rev3-ΔC | Wild-Type | rev3-ΔC | |
| 5′CC | 6 | 5 | 5 | 11 | 5 | 13 |
| 5′CT | 1 | 5 | 2 | 3 | 3 | 1 |
| 5′TC | 1 | 3 | 5 | 5 | 5 | 12 |
| 5′TT | 17 | 14 | 19 | 13 | 17 | 7 # |
| OVL | 9 | 15 | 6 | 11 | 8 | 14 |
| Total in DP sites (%) | 34 (85%) | 42 (93%) | 37 (84%) | 43 (86%) | 38 (81%) | 47 (94%) |
| Non-DP | 6 | 3 | 7 | 5 | 9 | 3 |
| Total changes | 40 | 45 | 44 | 48 | 47 | 50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siebler, H.M.; Cui, J.; Hill, S.E.; Pavlov, Y.I. DNA Polymerase ζ without the C-Terminus of Catalytic Subunit Rev3 Retains Characteristic Activity, but Alters Mutation Specificity of Ultraviolet Radiation in Yeast. Genes 2022, 13, 1576. https://doi.org/10.3390/genes13091576
Siebler HM, Cui J, Hill SE, Pavlov YI. DNA Polymerase ζ without the C-Terminus of Catalytic Subunit Rev3 Retains Characteristic Activity, but Alters Mutation Specificity of Ultraviolet Radiation in Yeast. Genes. 2022; 13(9):1576. https://doi.org/10.3390/genes13091576
Chicago/Turabian StyleSiebler, Hollie M., Jian Cui, Sarah E. Hill, and Youri I. Pavlov. 2022. "DNA Polymerase ζ without the C-Terminus of Catalytic Subunit Rev3 Retains Characteristic Activity, but Alters Mutation Specificity of Ultraviolet Radiation in Yeast" Genes 13, no. 9: 1576. https://doi.org/10.3390/genes13091576
APA StyleSiebler, H. M., Cui, J., Hill, S. E., & Pavlov, Y. I. (2022). DNA Polymerase ζ without the C-Terminus of Catalytic Subunit Rev3 Retains Characteristic Activity, but Alters Mutation Specificity of Ultraviolet Radiation in Yeast. Genes, 13(9), 1576. https://doi.org/10.3390/genes13091576