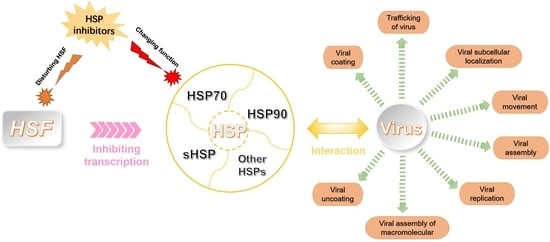
Mode of Action of Heat Shock Protein (HSP) Inhibitors against Viruses through Host HSP and Virus Interactions
Abstract
:1. Introduction
2. HSPs Are Important Molecular Chaperones
3. HSFs Can Promote the Transcription of HSP Genes
4. HSPs Participate in Different Viral Functions
4.1. HSPs Interact with Viral Proteins to Change the Activation Status
4.2. HSPs Are Involved in the Assembly of Macromolecular Protein Complexes
4.3. HSPs Are Involved with the Subcellular Localization of Virus Components
4.4. HSPs Are Involved in Viral Replication
4.5. HSPs Are Involved in Viral Translation
4.6. HSPs Are Involved in Viral Infection
4.7. HSPs Are Involved in Viral Movement
4.8. HSPs Are Involved with the Assembly of Viral Particles
4.9. HSPs Are Involved with the Disassembly of Viral Particles
4.10. HSPs Are Involved in Viral Transport
5. Viral Infection Can Change HSPs from Host and Virus
5.1. Human or Animal Viral Infection Can Change the Intracellular Distribution of HSPs from Mammal Cells
5.2. Viral Infection Can Enhance the Amount of HSPs in Mammal Cells
5.3. Viral Infection Can Enhance the Amount of HSPs in Plant Cells
5.4. Viral Infection Can Enhance the Amount of HSPs Encoded by Itself
5.5. The Mechanism of HSP Transcription Is Triggered by Viral Infection
6. HSP Inhibitors Can Downregulate HSPs at Many Levels
6.1. Quercetin Can Decrease the Expression Level of HSP70
6.2. Quercetin Regulates HSF at Many Levels
6.3. Flavonoid Compounds with Structural Similarity to Quercetin Can Decrease the Expression Level of HSPs or Change the Conformation of HSPs
6.4. Flavonoids with Structure Similar to Quercetin Regulate HSFs
6.5. KNK437 Can Decrease the mRNA Level of HSPs
6.6. Inhibitors PES and GA Can Disrupt the Interaction between HSP and Its Client Protein
6.7. Sodium Salicylate Changes the Expression of HSPs by Regulating HSFs
6.8. Some Potential Inhibitors of HSP90
6.9. Dihydromyricetin-Based HSP70 Inhibitors
6.10. Rhodacyanine-Based HSP70 Inhibitors
6.11. Chromone Analogs Possessing Thiiran-2-Ylmethoxy or Oxyran-2-Ylmethoxy Substituents-Based sHsp Inhibitors
7. The Antiviral Activity of HSP Inhibitors Indicated That These Compounds Can Regulate Viral Multiplication
7.1. Quercetin Possesses Potent Antiviral Activity
7.2. Quercetin Derivatives Exhibit Marked Antiviral Activity
7.3. Quercetin Analogs Exhibit Prominent Antiviral Activity
8. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alam, S.B.; Rochon, D. Cucumber necrosis virus recruits cellular heat shock protein 70 homologs at several stages of infection. J. Virol. 2015, 90, 3302–3317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.K.; Kim, R.; Kim, S.H. Crystal structure of a small heat-shock protein. Nature 1998, 394, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Young, J.C.; Agashe, V.R.; Siegers, K.; Hartl, F.U. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004, 5, 781–791. [Google Scholar] [CrossRef]
- Frydman, J. Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu. Rev. Biochem. 2001, 70, 603–647. [Google Scholar] [CrossRef] [PubMed]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Lindquist, S.; Craig, E.A. The heat-shock proteins. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef] [PubMed]
- Parsell, D.A.; Lindquist, S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993, 27, 437–496. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P. Recruitment of Hsp70 chaperones: A crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 2005, 153, 1–46. [Google Scholar] [CrossRef]
- Chen, Z.R.; Zhou, T.; Wu, X.H.; Hong, Y.G.; Fan, Z.F.; Li, H.F. Influence of cytoplasmic heat shock protein 70 on viral infection of Nicotiana benthamiana. Mol. Plant Pathol. 2008, 9, 809–817. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, J.; Sheng, Y.; Xiao, Y.F.; Zhang, Y.J.; Bai, L.X.; Tan, Y.; Xiao, L.B.; Xu, G.C. Identification of heat shock cognate protein 70 gene (Alhsc70) of Apolygus lucorum and its expression in response to different temperature and pesticide stresses. Insect Sci. 2016, 23, 37–49. [Google Scholar] [CrossRef]
- Chen, B.; Piel, W.; Gui, L.; Bruford, E.; Monteiro, A. The HSP90 family of genes in the human genome: Insights into their divergence and evolution. Genomics 2005, 86, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Mimnaugh, E.G.; Chavany, C.; Neckers, L. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J. Biol. Chem. 1996, 271, 22796–22801. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.; Sepp-Lorenzino, L.; Nimmesgern, E.; Ouerfelli, O.; Danishefsky, S.; Rosen, N.; Hartl, F.U. Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc. Natl. Acad. Sci. USA 1996, 93, 14536–14541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearl, L.H.; Prodromou, C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 2006, 75, 271–294. [Google Scholar] [CrossRef] [PubMed]
- Krukenberg, K.A.; Street, T.O.; Lavery, L.A.; Agard, D.A. Conformational dynamics of the molecular chaperone Hsp90. Q. Rev. Biophys. 2011, 44, 229–255. [Google Scholar] [CrossRef] [Green Version]
- Xiang, J.; Chen, X.; Hu, W.; Xiang, Y.; Yan, M.; Wang, J. Overexpressing heat-shock protein OsHSP50.2 improves drought tolerance in rice. Plant Cell Rep. 2018, 37, 1585–1595. [Google Scholar] [CrossRef]
- Samakovli, D.; Ticha, T.; Vavrdova, T.; Zavorkova, N.; Pecinka, A.; Ovecka, M.; Samaj, J. HEAT SHOCK PROTEIN 90 proteins and YODA regulate main body axis formation during early embryogenesis. Plant Physiol. 2021, 186, 1526–1544. [Google Scholar] [CrossRef]
- Zolkiewski, M.; Zhang, T.; Nagy, M. Aggregate reactivation mediated by the Hsp100 chaperones. Arch. Biochem. Biophys. 2012, 520, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Gurley, W.B. HSP101: A key component for the acquisition of thermotolerance in plants. Plant Cell 2000, 12, 457–460. [Google Scholar] [CrossRef] [Green Version]
- Qin, F.; Yu, B.; Li, W. Heat shock protein 101 (HSP101) promotes flowering under nonstress conditions. Plant Physiol. 2021, 186, 407–419. [Google Scholar] [CrossRef]
- Yu, C.; Leung, S.K.P.; Zhang, W.; Lai, L.T.F.; Chan, Y.K.; Wong, M.C.; Benlekbir, S.; Cui, Y.; Jiang, L.; Lau, W.C.Y. Structural basis of substrate recognition and thermal protection by a small heat shock protein. Nat. Commun. 2021, 12, 3007. [Google Scholar] [CrossRef] [PubMed]
- Kimmins, S.; MacRae, T.H. Maturation of steroid receptors: An example of functional cooperation among molecular chaperones and their associated proteins. Cell Stress Chaperon. 2000, 5, 76–86. [Google Scholar] [CrossRef]
- Pratt, W.B.; Toft, D.O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 2003, 228, 111–133. [Google Scholar] [CrossRef] [PubMed]
- Pearl, L.H.; Prodromou, C.; Workman, P. The Hsp90 molecular chaperone: An open and shut case for treatment. Biochem. J. 2008, 410, 439–453. [Google Scholar] [CrossRef] [Green Version]
- Dickey, C.A.; Koren, J.; Zhang, Y.J.; Xu, Y.F.; Jinwal, U.K.; Birnbaum, M.J.; Monks, B.; Sun, M.; Cheng, J.Q.; Patterson, C.; et al. Akt and CHIP coregulate tau degradation through coordinated interactions. Proc. Natl. Acad. Sci. USA 2008, 105, 3622–3627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murata, S.; Chiba, T.; Tanaka, K. CHIP: A quality-control E3 ligase collaborating with molecular chaperones. Int. J. Biochem. Cell Biol. 2003, 35, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Peteranderl, R.; Rabenstein, M.; Shin, Y.K.; Liu, C.W.; Wemmer, D.E.; King, D.S.; Nelson, H.C. Biochemical and biophysical characterization of the trimerization domain from the heat shock transcription factor. Biochemistry 1999, 38, 3559–3569. [Google Scholar] [CrossRef]
- Goldenberg, C.J.; Luo, Y.; Fenna, M.; Baler, R.; Weinmann, R.; Voellmy, R. Purified human factor activates heat shock promoter in a HeLa cell-free transcription system. J. Biol. Chem. 1988, 263, 19734–19739. [Google Scholar] [CrossRef]
- Susan, L. The heat-shock response. Annu. Rev. Biochem. 1986, 55, 1151–1191. [Google Scholar] [CrossRef]
- Phillips, B.; Abravaya, K.; Morimoto, R.I. Analysis of the specificity and mechanism of transcriptional activation of the human hsp70 gene during infection by DNA viruses. J. Virol. 1991, 65, 5680–5692. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.J.; Kingston, R.E.; Morimoto, R.I. Human HSP70 promoter contains at least two distinct regulatory domains. Proc. Natl. Acad. Sci. USA 1986, 83, 629–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larson, J.S.; Schuetz, T.J.; Kingston, R.E. Activation in vitro of sequence-specific DNA binding by a human regulatory factor. Nature 1988, 335, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Sorger, P.K.; Lewis, M.J.; Pelham, H.R. Heat shock factor is regulated differently in yeast and HeLa cells. Nature 1987, 329, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Nagai, N.; Nakai, A.; Nagata, K. Quercetin suppresses heat shock response by down regulation of HSF1. Biochem. Biophys. Res. Commun. 1995, 208, 1099–1105. [Google Scholar] [CrossRef]
- Zimarino, V.; Wu, C. Induction of sequence-specific binding of Drosophila heat shock activator protein without protein synthesis. Nature 1987, 327, 727–730. [Google Scholar] [CrossRef]
- Hosokawa, N.; Hirayoshi, K.; Nakai, A.; Hosokawa, Y.; Marui, N.; Yoshida, M.; Sakai, T.; Nishino, H.; Aoike, A.; Kawai, K.; et al. Flavonoids inhibit the expression of heat shock proteins. Cell Struct. Funct. 1990, 15, 393–401. [Google Scholar] [CrossRef] [Green Version]
- Mosser, D.D.; Theodorakis, N.G.; Morimoto, R.I. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol. Cell. Biol. 1988, 8, 4736–4744. [Google Scholar] [CrossRef] [Green Version]
- Qi, H.; Chen, X.; Luo, S.; Fan, H.; Guo, J.; Zhang, X.; Ke, Y.; Yang, P.; Yu, F. Genome-wide identification and characterization of heat shock protein 20 genes in maize. Life-Basel 2022, 12, 1397. [Google Scholar] [CrossRef]
- Sun, X.; Huang, N.; Li, X.; Zhu, J.; Bian, X.; Li, H.; Wang, L.; Hu, Q.; Luo, H. A chloroplast heat shock protein modulates growth and abiotic stress response in creeping bentgrass. Plant Cell Environ. 2021, 44, 1769–1787. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, Y.; Tao, X.; Wang, J.Z.; Cheng, H.Y.; Yang, H.; Ma, X.R. Heat shock factor genes of tall fescue and perennial ryegrass in response to temperature stress by RNA-Seq analysis. Front. Plant Sci. 2015, 6, 1226. [Google Scholar] [CrossRef] [Green Version]
- Bharti, K.; Von, K.P.; Bharti, S.; Kumar, P.; Tintschl-Korbitzer, A.; Treuter, E.; Nover, L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 2004, 16, 1521–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. BBA-Gene Regul. Mech. 2012, 1819, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Ahlquist, P.; Noueiry, A.O.; Lee, W.M.; Kushner, D.B.; Dye, B.T. Host factors in positive-strand RNA virus genome replication. J. Virol. 2003, 77, 8181–8186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kao, C.C.; Singh, P.; Ecker, D.J. De novo initiation of viral RNA-dependent RNA synthesis. Virology 2001, 287, 251–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basta, S.; Stoessel, R.; Basler, M.; van den Broek, M.; Groettrup, M. Cross-presentation of the long-lived lymphocytic choriomeningitis virus nucleoprotein does not require neosynthesis and is enhanced via heat shock proteins. J. Immunol. 2005, 175, 796–805. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Xie, E.Y.; Guo, H.Z.; Sun, Q.; Liuli, H.; Wang, Y.M.; Li, Q.; Xia, Q.Y. Heat shock protein 19.9 (Hsp19.9) from Bombyx mori is involved in host protection against viral infection. Dev. Comp. Immunol. 2021, 114, 103790. [Google Scholar] [CrossRef]
- Lin, T.W.; Lo, C.W.; Lai, S.Y.; Fan, R.J.; Lo, C.J.; Chou, Y.M.; Thiruvengadam, R.; Wang, A.H.; Wang, M.Y. Chicken heat shock protein 90 is a component of the putative cellular receptor complex of infectious bursal disease virus. J. Virol. 2007, 81, 8730–8741. [Google Scholar] [CrossRef] [Green Version]
- Connor, J.H.; McKenzie, M.O.; Parks, G.D.; Lyles, D.S. Antiviral activity and RNA polymerase degradation following Hsp90 inhibition in a range of negative strand viruses. Virology 2007, 362, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Karasev, A.V.; Kashina, A.S.; Gelfand, V.I.; Dolja, V.V. HSP70-related 65 kDa protein of beet yellows closterovirus is a microtubule-binding protein. FEBS Lett. 1992, 304, 12–14. [Google Scholar] [CrossRef] [Green Version]
- Alzhanova, D.V.; Napuli, A.J.; Creamer, R.; Dolja, V.V. Cell-to-cell movement and assembly of a plant closterovirus: Roles for the capsid proteins and Hsp70 homolog. EMBO J. 2001, 20, 6997–7007. [Google Scholar] [CrossRef] [Green Version]
- Zhu, P.; Lv, C.; Fang, C.; Peng, X.; Sheng, H.; Xiao, P.; Ojha, N.K.; Yan, Y.; Liao, M.; Zhou, J. Heat shock protein member 8 is an attachment factor for infectious bronchitis virus. Front. Microbiol. 2020, 11, 1630. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Ryu, J.; Park, J.E.; Hong, E.J.; Shin, H.J. Heat shock protein 70 could enhance porcine epidemic diarrhoea virus replication by interacting with membrane proteins. Vet. Res. 2021, 52, 138. [Google Scholar] [CrossRef]
- Smith, D.R.; McCarthy, S.; Chrovian, A.; Olinger, G.; Stossel, A.; Geisbert, T.W.; Hensley, L.E.; Connor, J.H. Inhibition of heat-shock protein 90 reduces Ebola virus replication. Antiviral Res. 2010, 87, 187–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabrera-Hernandez, A.; Thepparit, C.; Suksanpaisan, L.; Smith, D.R. Dengue virus entry into liver (HepG2) cells is independent of hsp90 and hsp70. J. Med. Virol. 2007, 79, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Ujino, S.; Yamaguchi, S.; Shimotohno, K.; Takaku, H. Heat-shock protein 90 is essential for stabilization of the hepatitis C virus nonstructural protein NS3. J. Biol. Chem. 2009, 284, 6841–6846. [Google Scholar] [CrossRef] [Green Version]
- Waxman, L.; Whitney, M.; Pollok, B.A.; Kuo, L.C.; Darke, P.L. Host cell factor requirement for hepatitis C virus enzyme maturation. Proc. Natl. Acad. Sci. USA 2001, 98, 13931–13935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Li, X.; Qian, Y.; Liu, C.; Huang, X.; Fu, M. Heat shock protein 90 inhibitors suppress pyroptosis in THP-1 cells. Biochem. J. 2020, 477, 3923–3934. [Google Scholar] [CrossRef]
- Okamoto, T.; Nishimura, Y.; Ichimura, T.; Suzuki, K.; Miyamura, T.; Suzuki, T.; Moriishi, K.; Matsuura, Y. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 2006, 25, 5015–5025. [Google Scholar] [CrossRef]
- Taguwa, S.; Kambara, H.; Omori, H.; Tani, H.; Abe, T.; Mori, Y.; Suzuki, T.; Yoshimori, T.; Moriishi, K.; Matsuura, Y. Cochaperone activity of human butyrate-induced transcript 1 facilitates hepatitis C virus replication through an Hsp90-dependent pathway. J. Virol. 2009, 83, 10427–10436. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, T.; Omori, H.; Kaname, Y.; Abe, T.; Nishimura, Y.; Suzuki, T.; Miyamura, T.; Yoshimori, T.; Moriishi, K.; Matsuura, Y. A single-amino-acid mutation in hepatitis C virus NS5A disrupting FKBP8 interaction impairs viral replication. J. Virol. 2008, 82, 3480–3489. [Google Scholar] [CrossRef] [Green Version]
- Taguwa, S.; Okamoto, T.; Abe, T.; Mori, Y.; Suzuki, T.; Moriishi, K.; Matsuura, Y. Human butyrate-induced transcript 1 interacts with hepatitis C virus NS5A and regulates viral replication. J. Virol. 2008, 82, 2631–2641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.J.; Chen, Y.H.; Chow, L.P.; Tsai, Y.H.; Chen, P.H.; Huang, C.Y.; Chen, W.T.; Hwang, L.H. Heat shock protein 72 is associated with the hepatitis C virus replicase complex and enhances viral RNA replication. J. Biol. Chem. 2010, 285, 28183–28190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Chen, Z.; Zhang, B.; Miao, H.; Zohaib, A.; Xu, Q.; Chen, H.; Cao, S. Heat shock protein 70 is associated with replicase complex of Japanese encephalitis virus and positively regulates viral genome replication. PLoS ONE 2013, 8, e75188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, G.; Han, K.; Huang, X.; Zhang, L.; Liu, Q.; Yang, J.; Liu, Y.; Li, Y.; Zhao, D. Heat shock protein 70 (HSP70) plays important role in tembusu virus infection. Vet. Microbiol. 2022, 267, 109377. [Google Scholar] [CrossRef]
- Pujhari, S.; Brustolin, M.; Macias, V.M.; Nissly, R.H.; Nomura, M.; Nomura, S.V.; Rasgon, J.L. Heat shock protein 70 (Hsp70) mediates Zika virus entry, replication, and egress from host cells. Emerg. Microbes Infect. 2019, 8, 8–16. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Toft, D.O.; Seeger, C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997, 16, 59–68. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Anselmo, D. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: Posttranslational activation by Hsp90. J. Virol. 2000, 74, 11447–11455. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Grammatikakis, N.; Hu, J. Role of p50/CDC37 in hepadnavirus assembly and replication. J. Biol. Chem. 2002, 277, 24361–24367. [Google Scholar] [CrossRef] [Green Version]
- Beck, J.; Nassal, M. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J. Biol. Chem. 2003, 278, 36128–36138. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Flores, D.; Toft, D.; Wang, X.; Nguyen, D. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J. Virol. 2004, 78, 13122–13131. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Seeger, C. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 1996, 93, 1060–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Z.Z.; Miao, J.; Zhao, M.; Tang, M.; Yeo, A.E.; Yu, H.; Zhang, J.; Xia, N.S. Role of heat-shock protein 90 in hepatitis E virus capsid trafficking. J. Gen. Virol. 2010, 91, 1728–1736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Barlow, E.A.; Ma, S.; Hagemeier, S.R.; Duellman, S.J.; Burgess, R.R.; Tellam, J.; Khanna, R.; Kenney, S.C. Hsp90 inhibitors block outgrowth of EBV-infected malignant cells in vitro and in vivo through an EBNA1-dependent mechanism. Proc. Natl. Acad. Sci. USA 2010, 107, 3146–3151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, Y.K.; Park, C.H.; Kim, K.Y.; Li, Y.C.; Kim, J.; Kim, Y.A.; Paik, J.H.; Park, B.K.; Kim, C.W.; Kim, Y.N. The heat-shock protein 90 inhibitor, geldanamycin, induces apoptotic cell death in Epstein-Barr virus-positive NK/T-cell lymphoma by Akt down-regulation. J. Pathol. 2007, 213, 170–179. [Google Scholar] [CrossRef]
- Kotsiopriftis, M.; Tanner, J.E.; Alfieri, C. Heat shock protein 90 expression in Epstein-Barr virus infected B cells promotes gammadelta T-cell proliferation in vitro. J. Virol. 2005, 79, 7255–7261. [Google Scholar] [CrossRef] [Green Version]
- Basha, W.; Kitagawa, R.; Uhara, M.; Imazu, H.; Uechi, K.; Tanaka, J. Geldanamycin, a potent and specific inhibitor of Hsp90, inhibits gene expression and replication of human cytomegalovirus. Antivir. Chem. Chemother. 2005, 16, 135–146. [Google Scholar] [CrossRef]
- Burch, A.D.; Weller, S.K. Herpes simplex virus type 1 DNA polymerase requires the mammalian chaperone hsp90 for proper localization to the nucleus. J. Virol. 2005, 79, 10740–10749. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.H.; Tao, P.Z.; Liu, Y.Z.; Jiang, J.D. Geldanamycin, a ligand of heat shock protein 90, inhibits the replication of herpes simplex virus type 1 in vitro. Antimicrob. Agents Chemother. 2004, 48, 867–872. [Google Scholar] [CrossRef] [Green Version]
- Kyratsous, C.A.; Silverstein, S.J. The co-chaperone BAG3 regulates Herpes Simplex Virus replication. Proc. Natl. Acad. Sci. USA 2008, 105, 20912–20917. [Google Scholar] [CrossRef] [Green Version]
- Wen, K.W.; Damania, B. Hsp90 and Hsp40/Erdj3 are required for the expression and anti-apoptotic function of KSHV K1. Oncogene 2010, 29, 3532–3544. [Google Scholar] [CrossRef] [Green Version]
- Field, N.; Low, W.; Daniels, M.; Howell, S.; Daviet, L.; Boshoff, C.; Collins, M. KSHV vFLIP binds to IKK-gamma to activate IKK. J. Cell Sci. 2003, 116, 3721–3728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyratsous, C.A.; Silverstein, S.J. BAG3, a host cochaperone, facilitates varicella-zoster virus replication. J. Virol. 2007, 81, 7491–7503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castorena, K.M.; Weeks, S.A.; Stapleford, K.A.; Cadwallader, A.M.; Miller, D.J. A functional heat shock protein 90 chaperone is essential for efficient flock house virus RNA polymerase synthesis in Drosophila cells. J. Virol. 2007, 81, 8412–8420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kampmueller, K.M.; Miller, D.J. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J. Virol. 2005, 79, 6827–6837. [Google Scholar] [CrossRef] [Green Version]
- Weeks, S.A.; Miller, D.J. The heat shock protein 70 cochaperone YDJ1 is required for efficient membrane-specific flock house virus RNA replication complex assembly and function in Saccharomyces cerevisiae. J. Virol. 2008, 82, 2004–2012. [Google Scholar] [CrossRef] [Green Version]
- Momose, F.; Naito, T.; Yano, K.; Sugimoto, S.; Morikawa, Y.; Nagata, K. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 2002, 277, 45306–45314. [Google Scholar] [CrossRef] [Green Version]
- Naito, T.; Momose, F.; Kawaguchi, A.; Nagata, K. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 2007, 81, 1339–1349. [Google Scholar] [CrossRef] [Green Version]
- Cao, M.M.; Wei, C.D.; Zhao, L.L.; Wang, J.F.; Jia, Q.N.; Wang, X.; Jin, Q.; Deng, T. DnaJA1/Hsp40 is co-opted by influenza A virus to enhance its viral RNA polymerase activity. J. Virol. 2014, 88, 14078–14089. [Google Scholar] [CrossRef] [Green Version]
- Chase, G.; Deng, T.; Fodor, E.; Leung, B.W.; Mayer, D.; Schwemmle, M.; Brownlee, G. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology 2008, 377, 431–439. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Erlichman, C.; McDonald, C.J.; Ingle, J.N.; Zollman, P.; Iankov, I.; Russell, S.J.; Galanis, E. Heat shock protein inhibitors increase the efficacy of measles virotherapy. Gene Ther. 2008, 15, 1024–1034. [Google Scholar] [CrossRef] [Green Version]
- Katoh, H.; Kubota, T.; Nakatsu, Y.; Tahara, M.; Kidokoro, M.; Takeda, M. Heat shock protein 90 ensures efficient mumps virus replication by assisting with viral polymerase complex formation. J. Virol. 2017, 91, e02220-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Shi, H.; Qi, R.; Sun, S.; Tang, Y.; Zhang, B.; Wang, C. Hsp90 regulates activation of interferon regulatory factor 3 and TBK-1 stabilization in Sendai virus-infected cells. Mol. Biol. Cell 2006, 17, 1461–1471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geller, R.; Vignuzzi, M.; Andino, R.; Frydman, J. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Gene. Dev. 2007, 21, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Lin, Z.; Wang, C.; Li, Y.; Xia, Y.; Zhao, M.; Hua, L.; Chen, Y.; Guo, M.; Zhu, B. Heat shock protein 70 as a supplementary receptor facilitates enterovirus 71 infections in vitro. Microb. Pathog. 2019, 128, 106–111. [Google Scholar] [CrossRef]
- Miyata, Y.; Yahara, I. p53-independent association between SV40 large T antigen and the major cytosolic heat shock protein, HSP90. Oncogene 2000, 19, 1477–1484. [Google Scholar] [CrossRef] [Green Version]
- Dufresne, P.J.; Thivierge, K.; Cotton, S.; Beauchemin, C.; Ide, C.; Ubalijoro, E.; Laliberte, J.F.; Fortin, M.G. Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 2008, 374, 217–227. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Wu, B.; Jiang, L.; Zhang, M.; Lu, Y.; Wang, S.; Yan, F.; Xin, X. Triticum aestivum heat shock protein 23.6 interacts with the coat protein of wheat yellow mosaic virus. Virus Genes 2019, 55, 209–217. [Google Scholar] [CrossRef]
- Hung, J.J.; Chung, C.S.; Chang, W. Molecular chaperone Hsp90 is important for vaccinia virus growth in cells. J. Virol. 2002, 76, 1379–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jindal, S.; Young, R.A. Vaccinia virus infection induces a stress response that leads to association of Hsp70 with viral proteins. J. Virol. 1992, 66, 5357–5362. [Google Scholar] [CrossRef] [Green Version]
- Rullen, C.B. HIV-1 auxiliary proteins: Making connections in a dying cell. Cell 1998, 93, 685–692. [Google Scholar] [CrossRef] [Green Version]
- O’Keeffe, B.; Fong, Y.; Chen, D.; Zhou, S.; Zhou, Q. Requirement for a kinase-specific chaperone pathway in the production of a Cdk9/cyclin T1 heterodimer responsible for P-TEFb-mediated tat stimulation of HIV-1 transcription. J. Biol. Chem. 2000, 275, 279–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakami, H.; Tomita, M.; Okudaira, T.; Ishikawa, C.; Matsuda, T.; Tanaka, Y.; Nakazato, T.; Taira, N.; Ohshiro, K.; Mori, N. Inhibition of heat shock protein-90 modulates multiple functions required for survival of human T-cell leukemia virus type I-infected T-cell lines and adult T-cell leukemia cells. Int. J. Cancer 2007, 120, 1811–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leone, G.; Coffey, M.C.; Gilmore, R.; Duncan, R.; Maybaum, L.; Lee, P.W. C-terminal trimerization, but not N-terminal trimerization, of the reovirus cell attachment protein is a posttranslational and Hsp70/ATP-dependent process. J. Biol. Chem. 1996, 271, 8466–8471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, D.; Chattopadhyay, S.; Bagchi, P.; Halder, U.C.; Nandi, S.; Mukherjee, A.; Kobayashi, N.; Taniguchi, K.; Chawla-Sarkar, M. Active participation of cellular chaperone Hsp90 in regulating the function of rotavirus nonstructural protein 3 (NSP3). J. Biol. Chem. 2011, 286, 20065–20077. [Google Scholar] [CrossRef] [Green Version]
- Qanungo, K.R.; Shaji, D.; Mathur, M.; Banerjee, A.K. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc. Natl. Acad. Sci. USA 2004, 101, 5952–5957. [Google Scholar] [CrossRef] [Green Version]
- Sakata, M.; Katoh, H.; Otsuki, N.; Okamoto, K.; Nakatsu, Y.; Lim, C.K.; Saijo, M.; Takeda, M.; Mori, Y. Heat shock protein 90 ensures the integrity of rubella virus p150 protein and supports viral replication. J. Virol. 2019, 93, e01142-19. [Google Scholar] [CrossRef]
- Lin, J.Y.; Nagy, P.D. Identification of novel host factors via conserved domain search: Cns1 cochaperone is a novel restriction factor of tombusvirus replication in yeast. J. Virol. 2013, 87, 12600–12610. [Google Scholar] [CrossRef] [Green Version]
- Pogany, J.; Stork, J.; Li, Z.; Nagy, P.D. In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc. Natl. Acad. Sci. USA 2008, 105, 19956–19961. [Google Scholar] [CrossRef] [Green Version]
- Mine, A.; Hyodo, K.; Tajima, Y.; Kusumanegara, K.; Taniguchi, T.; Kaido, M.; Mise, K.; Taniguchi, H.; Okuno, T. Differential roles of Hsp70 and Hsp90 in the assembly of the replicase complex of a positive-strand RNA plant virus. J. Virol. 2012, 86, 12091–12104. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Xiang, C.Y.; Yang, J.; Chen, J.P.; Zhang, H.M. Interaction of HSP20 with a viral RdRp changes its sub-cellular localization and distribution pattern in plants. Sci. Rep. 2015, 5, 14016. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Lu, Y.; Li, K.; Lin, L.; Zheng, H.; Yan, F.; Chen, J. Heat shock protein 70 is necessary for Rice stripe virus infection in plants. Mol. Plant Pathol. 2014, 15, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawai, E.T.; Butel, J.S. Association of a cellular heat shock protein with simian virus 40 large T antigen in transformed cells. J. Virol. 1989, 63, 3961–3973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gannon, J.V.; Lane, D.P. p53 and DNA polymerase alpha compete for binding to SV40 T antigen. Nature 1987, 329, 456–458. [Google Scholar] [CrossRef] [PubMed]
- Pogany, J.; White, K.A.; Nagy, P.D. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 2005, 79, 4859–4869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagy, P.D.; Pogany, J. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 2006, 344, 211–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, K.A.; Nagy, P.D. Advances in the molecular biology of tombusviruses: Gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 2004, 78, 187–226. [Google Scholar] [CrossRef]
- Serva, S.; Nagy, P.D. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 2006, 80, 2162–2169. [Google Scholar] [CrossRef] [Green Version]
- Gorovits, R.; Moshe, A.; Ghanim, M.; Czosnek, H. Recruitment of the host plant heat shock protein 70 by Tomato yellow leaf curl virus coat protein is required for virus infection. PLoS ONE 2013, 8, e70280. [Google Scholar] [CrossRef]
- Wang, R.Y.; Stork, J.; Nagy, P.D. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J. Virol. 2009, 83, 3276–3287. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.Y.; Stork, J.; Pogany, J.; Nagy, P.D. A temperature sensitive mutant of heat shock protein 70 reveals an essential role during the early steps of tombusvirus replication. Virology 2009, 394, 28–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirsch, R.C.; Lavine, J.E.; Chang, L.J.; Varmus, H.E.; Ganem, D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as wel as for reverse transcription. Nature 1990, 344, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Toft, D.; Anselmo, D.; Wang, X. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J. Virol. 2002, 76, 269–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burch, A.D.; Weller, S.K. Nuclear sequestration of cellular chaperone and proteasomal machinery during herpes simplex virus type 1 infection. J. Virol. 2004, 78, 7175–7185. [Google Scholar] [CrossRef] [Green Version]
- Pogany, J.; Nagy, P.D. Activation of tomato bushy stunt virus RNA-dependent RNA polymerase by cellular heat shock protein 70 is enhanced by phospholipids in vitro. J. Virol. 2015, 89, 5714–5723. [Google Scholar] [CrossRef] [Green Version]
- Mathioudakis, M.M.; Rodriguez-Moreno, L.; Sempere, R.N.; Aranda, M.A.; Livieratos, I. Multifaceted capsid proteins: Multiple interactions suggest multiple roles for Pepino mosaic virus capsid protein. Mol. Plant Microbe Interact. 2014, 27, 1356–1369. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.W.; Hu, C.C.; Liou, M.R.; Chang, B.Y.; Tsai, C.H.; Meng, M.; Lin, N.S.; Hsu, Y.H. Hsp90 interacts specifically with viral RNA and differentially regulates replication initiation of Bamboo mosaic virus and associated satellite RNA. PLoS Pathog. 2012, 8, e1002726. [Google Scholar] [CrossRef] [Green Version]
- Harb, M.; Becker, M.M.; Vitour, D.; Baron, C.H.; Vende, P.; Brown, S.C.; Bolte, S.; Arold, S.T.; Poncet, D. Nuclear localization of cytoplasmic poly(A)-binding protein upon rotavirus infection involves the interaction of NSP3 with eIF4G and RoXaN. J. Virol. 2008, 82, 11283–11293. [Google Scholar] [CrossRef] [Green Version]
- Michel, Y.M.; Poncet, D.; Piron, M.; Kean, K.M.; Borman, A.M. Cap-Poly(A) synergy in mammalian cell-free extracts: Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 2000, 275, 32268–32276. [Google Scholar] [CrossRef] [Green Version]
- Vende, P.; Piron, M.; Castagne, N.; Poncet, D. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3’ end. J. Virol. 2000, 74, 7064–7071. [Google Scholar] [CrossRef] [Green Version]
- Dutta, D.; Bagchi, P.; Chatterjee, A.; Nayak, M.K.; Mukherjee, A.; Chattopadhyay, S.; Nagashima, S.; Kobayashi, N.; Komoto, S.; Taniguchi, K.; et al. The molecular chaperone heat shock protein-90 positively regulates rotavirus infection. Virology 2009, 391, 325–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsutsumi, S.; Neckers, L. Extracellular heat shock protein 90: A role for a molecular chaperone in cell motility and cancer metastasis. Cancer Sci. 2007, 98, 1536–1539. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.; Chavez-Salinas, S.; Medina, F.; del Angel, R.M. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J. Virol. 2005, 79, 4557–4567. [Google Scholar] [CrossRef] [Green Version]
- Tsan, M.F.; Gao, B. Heat shock proteins and immune system. J. Leukocyte Biol. 2009, 85, 905–910. [Google Scholar] [CrossRef]
- Chavez-Salinas, S.; Ceballos-Olvera, I.; Valle, J.R.; Medina, F.; del Angel, R.M. Heat shock effect upon dengue virus replication into U937 cells. Virus Res. 2008, 138, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Carrington, J.C.; Kasschau, K.D.; Mahajan, S.K.; Schaad, M.C. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 1996, 8, 1669–1681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traynor, P.; Young, B.M.; Ahlquist, P. Deletion analysis of brome mosaic virus 2a protein: Effects on RNA replication and systemic spread. J. Virol. 1991, 65, 2807–2815. [Google Scholar] [CrossRef] [Green Version]
- Gal-On, A.; Kaplan, I.; Roossinck, M.J.; Palukaitis, P. The kinetics of infection of zucchini squash by cucumber mosaic virus indicate a function for RNA 1 in virus movement. Virology 1994, 205, 280–289. [Google Scholar] [CrossRef]
- Holt, C.A.; Hodgson, R.A.; Coker, F.A.; Beachy, R.N.; Nelson, R.S. Characterization of the masked strain of tobacco mosaic virus: Identification of the region responsible for symptom attenuation by analysis of an infectious cDNA clone. Mol. Plant Microbe Interact. 1990, 3, 417–423. [Google Scholar] [CrossRef]
- Krenz, B.; Windeisen, V.; Wege, C.; Jeske, H.; Kleinow, T. A plastid-targeted heat shock cognate 70kDa protein interacts with the Abutilon mosaic virus movement protein. Virology 2010, 401, 6–17. [Google Scholar] [CrossRef] [Green Version]
- Wolf, S.; Deom, C.M.; Beachy, R.N.; Lucas, W.J. Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 1989, 246, 377–379. [Google Scholar] [CrossRef] [PubMed]
- Oparka, K.J.; Roberts, A.G.; Boevink, P.; Cruz, S.S.; Roberts, I.; Pradel, K.S.; Imlau, A.; Kotlizky, G.; Sauer, N.; Epel, B. Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 1999, 97, 743–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanlent, J.; Storms, M.; Vandermeer, F.; Wellink, J.; Goldbach, R. Tubular structures involved in movement of cowpea mosaic virus are also formed in infected cowpea protoplasts. J. Gen. Virol. 1991, 72, 2615–2623. [Google Scholar] [CrossRef]
- Perbal, M.C.; Thomas, C.L.; Maule, A.J. Cauliflower mosaic virus gene I product (P1) forms tubular structures which extend from the surface of infected protoplasts. Virology 1993, 195, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Agranovsky, A.A.; Boyko, V.P.; Karasev, A.V.; Koonin, E.V.; Dolja, V.V. Putative 65 kDa protein of beet yellows closterovirus is a homologue of HSP70 heat shock proteins. J. Mol. Biol. 1991, 217, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Fazeli, C.F.; Rezaian, M.A. Nucleotide sequence and organization of ten open reading frames in the genome of Grapevine leafroll-associated virus 1 and identification of three subgenomic RNAs. J. Gen. Virol. 2000, 81, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Prokhnevsky, A.I.; Peremyslov, V.V.; Napuli, A.J.; Dolja, V.V. Interaction between long-distance transport factor and Hsp70-related movement protein of Beet yellows virus. J. Virol. 2002, 76, 11003–11011. [Google Scholar] [CrossRef] [Green Version]
- Medina, V.; Peremyslov, V.V.; Hagiwara, Y.; Dolja, V.V. Subcellular localization of the HSP70-homolog encoded by beet yellows closterovirus. Virology 1999, 260, 173–181. [Google Scholar] [CrossRef] [Green Version]
- Peremyslov, V.V.; Hagiwara, Y.; Dolja, V.V. HSP70 homolog functions in cell-to-cell movement of a plant virus. Proc. Natl. Acad. Sci. USA 1999, 96, 14771–14776. [Google Scholar] [CrossRef] [Green Version]
- Geller, R.; Taguwa, S.; Frydman, J. Broad action of Hsp90 as a host chaperone required for viral replication. BBA-Mol. Cell Res. 2012, 1823, 698–706. [Google Scholar] [CrossRef] [Green Version]
- Cripe, T.P.; Delos, S.E.; Estes, P.A.; Garcea, R.L. In vivo and in vitro association of hsc70 with polyomavirus capsid proteins. J. Virol. 1995, 69, 7807–7813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, H.W.; Seo, J.P.; Jung, G. Heat shock protein 70 and heat shock protein 90 synergistically increase hepatitis B viral capsid assembly. Biochem. Biophys. Res. Commun. 2018, 503, 2892–2898. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, J.; Shuman, S. Novobiocin inhibits vaccinia virus replication by blocking virus assembly. Virology 1997, 235, 129–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekiguchi, J.; Stivers, J.T.; Mildvan, A.S.; Shuman, S. Mechanism of inhibition of vaccinia DNA topoisomerase by novobiocin and coumermycin. J. Biol. Chem. 1996, 271, 2313–2322. [Google Scholar] [CrossRef] [Green Version]
- Ivanovic, T.; Agosto, M.A.; Chandran, K.; Nibert, M.L. A role for molecular chaperone Hsc70 in reovirus outer capsid disassembly. J. Biol. Chem. 2007, 282, 12210–12219. [Google Scholar] [CrossRef] [Green Version]
- Gething, M.J.; Mccammon, K.; Sambrook, J. Expression of wild-type and mutant forms of influenza hemagglutinin: The role of folding in intracellular transport. Cell 1986, 46, 939–950. [Google Scholar] [CrossRef]
- Knarr, G.; Modrow, S.; Todd, A.; Gething, M.J.; Buchner, J. BiP-binding sequences in HIV gp160: Implications for the binding specificity of Bip. J. Biol. Chem. 1999, 274, 29850–29857. [Google Scholar] [CrossRef] [Green Version]
- Brown, G.; Rixon, H.W.; Steel, J.; McDonald, T.P.; Pitt, A.R.; Graham, S.; Sugrue, R.J. Evidence for an association between heat shock protein 70 and the respiratory syncytial virus polymerase complex within lipid-raft membranes during virus infection. Virology 2005, 338, 69–80. [Google Scholar] [CrossRef] [Green Version]
- LaThangue, N.B.; Shriver, K.; Dawson, C.; Chan, W.L. Herpes simplex virus infection causes the accumulation of a heat-shock protein. EMBO J. 1984, 3, 267–277. [Google Scholar] [CrossRef]
- Kao, H.T.; Nevins, J.R. Transcriptional activation and subsequent control of the human heat shock gene during adenovirus infection. Mol. Cell. Biol. 1983, 3, 2058–2065. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.J.; Hurst, H.C.; Jones, N.C.; Morimoto, R.I. The E1A 13S product of adenovirus 5 activates transcription of the cellular human HSP70 gene. Mol. Cell. Biol. 1986, 6, 2994–2999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathioudakis, M.M.; Veiga, R.; Ghita, M.; Tsikou, D.; Medina, V.; Canto, T.; Makris, A.M.; Livieratos, I.C. Pepino mosaic virus capsid protein interacts with a tomato heat shock protein cognate 70. Virus Res. 2012, 163, 28–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aparicio, F.; Thomas, C.L.; Lederer, C.; Niu, Y.; Wang, D.; Maule, A.J. Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol. 2005, 138, 529–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agranovsky, A.A.; Koonin, E.V.; Boyko, V.P.; Maiss, E.; Frotschl, R.; Lunina, N.A.; Atabekov, J.G. Beet yellows closterovirus: Complete genome structure and identification of a leader papain-like thiol protease. Virology 1994, 198, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Coutts, R.H.; Coffin, R.S. Beet pseudo-yellows virus is an authentic closterovirus. Virus Genes 1996, 13, 179–181. [Google Scholar] [CrossRef]
- Alwine, J.C. Transient gene expression control: Effects of transfected DNA stability and trans-activation by viral early proteins. Mol. Cell. Biol. 1985, 5, 1034–1042. [Google Scholar] [CrossRef] [Green Version]
- Berk, A.J.; Lee, F.; Harrison, T.; Williams, J.; Sharp, P.A. Pre-early adenovirus 5 gene product regulates synthesis of early viral messenger RNAs. Cell 1979, 17, 935–944. [Google Scholar] [CrossRef]
- Brady, J.; Bolen, J.B.; Radonovich, M.; Salzman, N.; Khoury, G. Stimulation of simian virus 40 late gene expression by simian virus 40 tumor antigen. Proc. Natl. Acad. Sci. USA 1984, 81, 2040–2044. [Google Scholar] [CrossRef] [Green Version]
- Brady, J.; Khoury, G. trans Activation of the simian virus 40 late transcription unit by T-antigen. Mol. Cell. Biol. 1985, 5, 1391–1399. [Google Scholar] [CrossRef] [Green Version]
- Brady, J.; Loeken, M.R.; Khoury, G. Interaction between two transcriptional control sequences required for tumor-antigen-mediated simian virus 40 late gene expression. Proc. Natl. Acad. Sci. USA 1985, 82, 7299–7303. [Google Scholar] [CrossRef] [Green Version]
- Gaynor, R.B.; Hillman, D.; Berk, A.J. Adenovirus early region 1A protein activates transcription of a nonviral gene introduced into mammalian cells by infection or transfection. Proc. Natl. Acad. Sci. USA 1984, 81, 1193–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, N.; Shenk, T. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc. Natl. Acad. Sci. USA 1979, 76, 3665–3669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, J.M.; Alwine, J.C. Activation of the SV40 late promoter: Direct effects of T antigen in the absence of viral DNA replication. Cell 1984, 36, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Keller, J.M.; Alwine, J.C. Analysis of an activatable promoter: Sequences in the simian virus 40 late promoter required for T-antigen-mediated trans activation. Mol. Cell. Biol. 1985, 5, 1859–1869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudray, P.; Tyndall, C.; Kamen, R.; Cuzin, F. The high affinity binding site on polyomavirus DNA for the viral large T protein. Nucleic Acids Res. 1981, 9, 5697–5710. [Google Scholar] [CrossRef] [Green Version]
- Tegtmeyer, P.; Andersen, B.; Shaw, S.B.; Wilson, V.G. Alternative interactions of the SV40 A protein with DNA. Virology 1981, 115, 75–87. [Google Scholar] [CrossRef]
- Kingston, R.E.; Cowie, A.; Morimoto, R.I.; Gwinn, K.A. Binding of polyomavirus large T antigen to the human hsp70 promoter is not required for trans activation. Mol. Cell. Biol. 1986, 6, 3180–3190. [Google Scholar] [CrossRef] [Green Version]
- Leung, W.C. Evidence for a herpes simplex virus-specific factor controlling the transcription of deoxypyrimidine kinase. J. Virol. 1978, 27, 269–274. [Google Scholar] [CrossRef] [Green Version]
- Cowie, A.; Kamen, R. Guanine nucleotide contacts within viral DNA sequences bound by polyomavirus large T antigen. J. Virol. 1986, 57, 505–514. [Google Scholar] [CrossRef] [Green Version]
- Pomerantz, B.J.; Hassell, J.A. Polyomavirus and simian virus 40 large T antigens bind to common DNA sequences. J. Virol. 1984, 49, 925–937. [Google Scholar] [CrossRef] [Green Version]
- Kelly, G.S. Quercetin. Monograph. Altern. Med. Rev. 2011, 16, 172–194. [Google Scholar] [PubMed]
- Larson, A.J.; Symons, J.D.; Jalili, T. Therapeutic potential of quercetin to decrease blood pressure: Review of efficacy and mechanisms. Adv. Nutr. 2012, 3, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Liu, Y.M.; Yang, S.; Song, B.A.; Xu, G.F.; Bhadury, P.S.; Jin, L.H.; Hu, D.Y.; Liu, F.; Xue, W.; et al. Studies on the chemical constituents and anticancer activity of Saxifraga stolonifera (L) Meeb. Bioorgan. Med. Chem. 2008, 16, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Dower, J.I.; Geleijnse, J.M.; Gijsbers, L.; Schalkwijk, C.; Kromhout, D.; Hollman, P.C. Supplementation of the pure flavonoids epicatechin and quercetin affects some biomarkers of endothelial dysfunction and inflammation in (pre)hypertensive adults: A randomized double-blind, placebo-controlled, crossover trial. J. Nutr. 2015, 145, 1459–1463. [Google Scholar] [CrossRef] [Green Version]
- Murakami, A.; Ashida, H.; Terao, J. Multitargeted cancer prevention by quercetin. Cancer Lett. 2008, 269, 315–325. [Google Scholar] [CrossRef]
- Carullo, G.; Cappello, A.R.; Frattaruolo, L.; Badolato, M.; Armentano, B.; Aiello, F. Quercetin and derivatives: Useful tools in inflammation and pain management. Future Med. Chem. 2017, 9, 79–93. [Google Scholar] [CrossRef]
- Cai, X.; Fang, Z.; Dou, J.; Yu, A.; Zhai, G. Bioavailability of quercetin: Problems and promises. Curr. Med. Chem. 2013, 20, 2572–2582. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Lu, K.; Zhou, S.; Zhang, S.; Xie, F.; Qi, B.; Li, Y. Development of an oil-in-water emulsion stabilized by a black bean protein-based nanocomplex for co-delivery of quercetin and perilla oil. LWT-Food Sci. Technol. 2015, 138, 110644. [Google Scholar] [CrossRef]
- Hussein, J.; El-Naggar, M.E. Synthesis of an environmentally quercetin nanoemulsion to ameliorate diabetic-induced cardiotoxicity. Biocatal. Agric. Biotechnol. 2021, 33, 101983. [Google Scholar] [CrossRef]
- Manca, M.L.; Lai, F.; Pireddu, R.; Valenti, D.; Schlich, M.; Pini, E.; Ailuno, G.; Fadda, A.M.; Sinico, C. Impact of nanosizing on dermal delivery and antioxidant activity of quercetin nanocrystals. J. Drug Delivery Sci. Technol. 2020, 55, 101482. [Google Scholar] [CrossRef]
- Storniolo, A.; Raciti, M.; Cucina, A.; Bizzarri, M.; Di Renzo, L. Quercetin affects Hsp70/IRE1α mediated protection from death induced by endoplasmic reticulum stress. Oxid. Med. Cell. Longevity 2015, 2015, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.F.; Nieh, S.; Jao, S.W.; Liu, C.L.; Wu, C.H.; Chang, Y.C.; Yang, C.Y.; Lin, Y.S. Quercetin suppresses drug-resistant spheres via the p38 MAPK-Hsp27 apoptotic pathway in oral cancer cells. PLoS ONE 2012, 7, e49275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Zhao, H.K.; Dong, Q.L.; Zhang, Y.Y.; Wang, Y.M.; Li, H.Y.; Xing, G.J.; Li, Q.Y.; Dong, Y.S. Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.). Front. Plant Sci. 2015, 6, 773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, R.K.; Oesterreich, S.; Lemieux, P.; Sarge, K.D.; Fuqua, S.A. Quercetin inhibits heat shock protein induction but not heat shock factor DNA-binding in human breast carcinoma cells. Biochem. Biophys. Res. Commun. 1997, 239, 851–856. [Google Scholar] [CrossRef]
- Kim, S.H.; Yeo, G.S.; Lim, Y.S.; Kang, C.D.; Kim, C.M.; Chung, B.S. Suppression of multidrug resistance via inhibition of heat shock factor by quercetin in MDR cells. Exp. Mol. Med. 1998, 30, 87–92. [Google Scholar] [CrossRef]
- Mosser, D.D.; Kotzbauer, P.T.; Sarge, K.D.; Morimoto, R.I. In vitro activation of heat shock transcription factor DNA-binding by calcium and biochemical conditions that affect protein conformation. Proc. Natl. Acad. Sci. USA 1990, 87, 3748–3752. [Google Scholar] [CrossRef] [Green Version]
- Hosokawa, N.; Hirayoshi, K.; Kudo, H.; Takechi, H.; Aoike, A.; Kawai, K.; Nagata, K. Inhibition of the activation of heat shock factor in vivo and in vitro by flavonoids. Mol. Cell. Biol. 1992, 12, 3490–3498. [Google Scholar] [CrossRef] [Green Version]
- Tran, P.L.; Kim, S.A.; Choi, H.S.; Yoon, J.H.; Ahn, S.G. Epigallocatechin-3-gallate suppresses the expression of HSP70 and HSP90 and exhibits anti-tumor activity in vitro and in vivo. BMC Cancer 2010, 10, 276. [Google Scholar] [CrossRef] [Green Version]
- Yin, Z.; Henry, E.C.; Gasiewicz, T.A. (-)-Epigallocatechin-3-gallate is a novel Hsp90 inhibitor. Biochemistry 2009, 48, 336–345. [Google Scholar] [CrossRef] [Green Version]
- Yokota, S.; Kitahara, M.; Nagata, K. Benzylidene lactam compound, KNK437, a novel inhibitor of acquisition of thermotolerance and heat shock protein induction in human colon carcinoma cells. Cancer Res. 2000, 60, 2942–2948. [Google Scholar] [CrossRef]
- Manwell, L.A.; Heikkila, J.J. Examination of KNK437- and quercetin-mediated inhibition of heat shock-induced heat shock protein gene expression in Xenopus laevis cultured cells. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 148, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Strom, E.; Sathe, S.; Komarov, P.G.; Chernova, O.B.; Pavlovska, I.; Shyshynova, I.; Bosykh, D.A.; Burdelya, L.G.; Macklis, R.M.; Skaliter, R.; et al. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat. Chem. Biol. 2006, 2, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Leu, J.I.; Pimkina, J.; Frank, A.; Murphy, M.E.; George, D.L. A small molecule inhibitor of inducible heat shock protein 70. Mol. Cell 2009, 36, 15–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeBoer, C.; Meulman, P.A.; Wnuk, R.J.; Peterson, D.H. Geldanamycin, a new antibiotic. J. Antibiot. 1970, 23, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Gorska, M.; Popowska, U.; Sielicka-Dudzin, A.; Kuban-Jankowska, A.; Sawczuk, W.; Knap, N.; Cicero, G.; Wozniak, F. Geldanamycin and its derivatives as Hsp90 inhibitors. Front. Biosci. Landmark 2012, 17, 2269–2277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stebbins, C.E.; Russo, A.A.; Schneider, C.; Rosen, N.; Hartl, F.U.; Pavletich, N.P. Crystal structure of an Hsp90-geldanamycin complex: Targeting of a protein chaperone by an antitumor agent. Cell 1997, 89, 239–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathore, A.P.; Haystead, T.; Das, P.K.; Merits, A.; Ng, M.L.; Vasudevan, S.G. Chikungunya virus nsP3 & nsP4 interacts with HSP-90 to promote virus replication: HSP-90 inhibitors reduce CHIKV infection and inflammation in vivo. Antivir. Res. 2014, 103, 7–16. [Google Scholar] [CrossRef]
- Jurivich, D.A.; Pachetti, C.; Qiu, L.; Welk, J.F. Salicylate triggers heat shock factor differently than heat. J. Biol. Chem. 1995, 270, 24489–24495. [Google Scholar] [CrossRef] [Green Version]
- Lamut, A.; Gjorgjieva, M.; Naesens, L.; Liekens, S.; Lillsunde, K.E.; Tammela, P.; Kikelj, D.; Tomasic, T. Anti-influenza virus activity of benzo[d]thiazoles that target heat shock protein 90. Bioorg. Chem. 2020, 98, 103733. [Google Scholar] [CrossRef]
- Oh, S.H.; Woo, J.K.; Yazici, Y.D.; Myers, J.N.; Kim, W.Y.; Jin, Q.; Hong, S.S.; Park, H.J.; Suh, Y.G.; Kim, K.W.; et al. Structural basis for depletion of heat shock protein 90 client proteins by deguelin. J. Natl. Cancer Inst. 2007, 99, 949–961. [Google Scholar] [CrossRef] [Green Version]
- Chang, D.J.; An, H.; Kim, K.S.; Kim, H.H.; Jung, J.; Lee, J.M.; Kim, N.J.; Han, Y.T.; Yun, H.; Lee, S.; et al. Design, synthesis, and biological evaluation of novel deguelin-based heat shock protein 90 (hsp90) inhibitors targeting proliferation and angiogenesis. J. Med. Chem. 2012, 55, 10863–10884. [Google Scholar] [CrossRef] [PubMed]
- Hastings, J.M.; Hadden, M.K.; Blagg, B.S.J. Synthesis and evaluation of derrubone and select analogues. J. Org. Chem. 2008, 73, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Brandt, G.E.; Galam, L.; Matts, R.L.; Blagg, B.S.J. Identification and initial sar of silybin: An hsp90 inhibitor. Bioorg. Med. Chem. Lett. 2011, 21, 2659–2664. [Google Scholar] [CrossRef]
- Bhadresha, K.; Upadhyay, V.; Kumar, S.P.; Pandya, P.; Jain, N.; Rawal, R.M. Computational investigation of ginkgetin and theaflavin as potential inhibitors of heat shock protein 90 (Hsp90). J. Biomol. Struct. Dyn. 2022, 40, 13675–13681. [Google Scholar] [CrossRef]
- Sun, B.; Tan, D.; Pan, D.; Baker, M.R.; Liang, Z.; Wang, Z.; Lei, J.; Liu, S.; Hu, C.Y.; Li, Q.X. Dihydromyricetin imbues antiadipogenic effects on 3T3-L1 cells via direct interactions with 78-kDa glucose-regulated protein. J. Nutr. 2021, 151, 1717–1725. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.S.; Kumar, V.; Lee, D.Y.; Chen, Y.; Wu, Y.C.; Gao, J.Y.; Chu, P.C. Development of novel rhodacyanine-based heat shock protein 70 inhibitors. Curr. Med. Chem. 2021, 28, 5431–5446. [Google Scholar] [CrossRef]
- Hwang, S.Y.; Kwak, S.Y.; Kwon, Y.; Lee, Y.S.; Na, Y. Synthesis and biological effect of chrom-4-one derivatives as functional inhibitors of heat shock protein 27. Eur. J. Med. Chem. 2017, 139, 892–900. [Google Scholar] [CrossRef]
- Kaul, T.N.; Elliott, M.M.D.; Ogra, P.L. Antiviral effect of flavonoids on human viruses. J. Med. Virol. 1985, 15, 71–79. [Google Scholar] [CrossRef]
- Gonzalez, O.; Fontanes, V.; Raychaudhuri, S.; Loo, R.; Loo, J.; Arumugaswami, V.; Sun, R.; Dasgupta, A.; French, S.W. The heat shock protein inhibitor Quercetin attenuates hepatitis C virus production. Hepatology 2009, 50, 1756–1764. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Sun, G.; Guo, W.; Huang, Y.; Sun, W.; Zhao, F.; Hu, K. Inhibition of hepatitis B virus replication by quercetin in human hepatoma cell lines. Virol. Sin. 2015, 30, 261–268. [Google Scholar] [CrossRef]
- Wu, W.; Li, R.; Li, X.; He, J.; Jiang, S.; Liu, S.; Yang, J. Quercetin as an antiviral agent inhibits influenza A virus (IAV) entry. Viruses 2016, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Gravina, H.D.; Tafuri, N.F.; Silva Junior, A.; Fietto, J.L.; Oliveira, T.T.; Diaz, M.A.; Almeida, M.R. In vitro assessment of the antiviral potential of trans-cinnamic acid, quercetin and morin against equid herpesvirus 1. Res. Vet. Sci. 2011, 91, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Mucsi, I.; Pragai, B.M. Inhibition of virus multiplication and alteration of cyclic AMP level in cell cultures by flavonoids. Experientia 1985, 41, 930–931. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xiao, S.; Liu, X.; Wang, L.; Ji, Q.; Mo, D.; Chen, Y. Inhibition of HSP70 reduces porcine reproductive and respiratory syndrome virus replication in vitro. BMC Microbiol. 2014, 14, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Narayanan, S.; Chang, K.O. Inhibition of influenza virus replication by plant-derived isoquercetin. Antiviral Res. 2010, 88, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Thapa, M.; Kim, Y.; Desper, J.; Chang, K.O.; Hua, D.H. Synthesis and antiviral activity of substituted quercetins. Bioorg. Med. Chem. Lett. 2012, 22, 353–356. [Google Scholar] [CrossRef] [Green Version]
- Karimova, E.; Baltina, L.; Spirikhin, L.; Gabbasov, T.; Orshanskaya, Y.; Zarubaev, V. Synthesis and antiviral activity of quercetin brominated derivatives. Nat. Prod. Commun. 2015, 10, 1565–1568. [Google Scholar] [CrossRef] [Green Version]
- Dayem, A.A.; Choi, H.Y.; Kim, Y.B.; Cho, S.G. Antiviral effect of methylated flavonol isorhamnetin against influenza. PLoS ONE 2015, 10, e0121610. [Google Scholar] [CrossRef] [Green Version]
- Song, J.M.; Lee, K.H.; Seong, B.L. Antiviral effect of catechins in green tea on influenza virus. Antiviral Res. 2005, 68, 66–74. [Google Scholar] [CrossRef]
- Ji, P.; Chen, C.; Hu, Y.; Zhan, Z.; Pan, W.; Li, R.; Li, E.; Ge, H.M.; Yang, G. Antiviral activity of Paulownia tomentosa against enterovirus 71 of hand, foot, and mouth disease. Biol. Pharm. Bull. 2015, 38, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.Y.; Li, M.L.; Huang, P.N.; Chien, K.Y.; Horng, J.T.; Shih, S.R. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J. Gen. Virol. 2008, 89, 2540–2549. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.Y.; Shih, S.R.; Pan, M.; Li, C.; Lue, C.F.; Stollar, V.; Li, M.L. hnRNP A1 interacts with the 5’ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J. Virol. 2009, 83, 6106–6114. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| Viral Family | Virus | Viral Protein | Molecular Chaperones and Other Host Factors | Action and Process | Source of MC | References |
|---|---|---|---|---|---|---|
| Arenaviridae | LCMV | Nucleoprotein | HSP90 | Antigen cross presentation | Human | [45] |
| Baculoviridae | BmNPV | Undetermined | HSP70 | Viral replication | Animal | [46] |
| Birnaviridae | IBDV | VP2 | HSP90 | Virus Internalization | Animal | [47] |
| Bunyaviridae | LACV | L protein | HSP90 | Protein stabilization | Human | [48] |
| Closteroviridae | BYV | Viral HSP70h | Microtubules protein and other proteins | Viral movement and assembly | Virus | [49,50] |
| Coronaviridae | IBV | Spike protein of IBV | HSPA8 | Viral entry | Animal | [51] |
| PEDV | membrane protein | HSP70 | Viral replication | Animal | [52] | |
| Filoviridae | Ebola virus | RdRp | HSP90 | Virus propagation | Human | [53] |
| Flaviviridae | DENV | Viral receptor | Bip | Virus Internalization | Human | [54] |
| HCV | NS3 Protease | HSP90 | Cleavage at NS2/3 junction, NS3 function | Human | [55,56,57] | |
| HCV | NS5A | FKBP8, hB-ind1, HSP90, and HSP72 | Replication complex formation/genome replication | Human | [58,59,60,61,62] | |
| JEV | NS3 and NS5 | HSP70 | Virus replication | Human | [63] | |
| TMUV | Capsid | HSP70 | Viral replication, assembly, and release | Animal | [64] | |
| ZIKV | Capsid and viral RNA | HSP70 | Viral entry, viral RNA production, and virion release | Humam | [65] | |
| Hepadnaviridae | DHBV | P protein | p23, Cdc37, HSP40, and HSC70 | Reverse transcriptase priming | Human | [66,67,68,69] |
| HBV | P protein | Hsp70, Hsp40, Hop, and P23 | Reverse transcriptase priming | Human | [66,70,71] | |
| Hepeviridae | HEV | Capsid | HSP90 | Intracellular transfer | Human | [72] |
| Herpesviridae | EBV | EBNA | HSP90 | Cell proliferation | Human | [73] |
| EBV | KH | PI90K | Apoptosis prevention | Human | [74,75] | |
| HCMV | MIE | PI3K | Expression of immediate early protein IE2 | Human | [76] | |
| HSV 1 | UL30 | HSP70 and BAG3 | Polymerase localization | Human | [77,78,79] | |
| HSV 2 | UL30 | HSP70 and BAG3 | Polymerase localization | Human | [77,78,79] | |
| KSHV | K1 | HSP40 | Apoptosis prevention | Human | [80] | |
| KSHV | v-FLIP | IKK and Cdc37 | Apoptosis prevention | Human | [81] | |
| VZV | ORF29F | HSP70 and BAG3 | Localization of ORF29 | Human | [82] | |
| Nodaviridae | FHV | Protein A | HSP90 | Replication complex formation | Animal | [83,84,85] |
| Orthomyxoviridae | Influenza A virus | PB1 and PB2 | HSP90 | Nuclear localization | Human | [86] |
| Influenza A virus | RdRp | HSP90 | Assembly and nuclear transport of viral RNA polymerase subunits | Human | [87] | |
| Influenza A virus | RdRp | HSP90 | Viral RNA polymerase complex formation | Human | [87] | |
| Influenza A virus | RdRp | HSP90 and DnaJA1 | RNA synthesis | Human | [88,89] | |
| Paramyxoviridae | HPIV 2/3 | L protein | HSP90 | Protein stabilization and replication | Human | [48] |
| MeV | MV-N | HSP70 | Enhanced oncolytic activity | Human | [90] | |
| MuV | Viral polymerase | HSP90 | Viral replication | Human | [91] | |
| SeV | - | TBK1, IRF3, and HSP90 | Innate immunity activation and signal transduction | Human and animal | [92] | |
| SV 5/41 | L protein | HSP90 | Human and animal | [48] | ||
| Picornaviridae | Coxsackievirus | P1 capsid protein | p23 | Cleavage of P1 into VP1, VP2, and VP3 | Human | [93] |
| Enterovirus 71 (EV71) | Viral particle | HSP70 | Initial binding of virus to host cells | Human | [94] | |
| Poliovirus | P1 capsid protein | p23 | Cleavage of P1 into VP1, VP2, and VP3 | Human | [93] | |
| Rhinovirus | P1 capsid protein | p23 | Cleavage of P1 into VP1, VP2, and VP3 | Human | [93] | |
| Polyomaviridae | SV 40 | LT | HSP90 | Stabilization of LT protein | Human and animal | [95] |
| Potyviridae | TuMV | RdRp | HSC70-3 and PABP | Viral replication | Plant | [96] |
| WYMV | Undetermined | HSP23.6 | Viral replication | Plant | [97] | |
| Poxviridae | VACV | 4a core protein | HSP70 | Capsid assembly/virus gene expression | Human and animal | [98,99] |
| Retrovirus | HIV 1 | tat | HSP90 | Transcription/cell survival | Human | [100,101] |
| HTLV | tat | HSP90 | Transcription | Human | [101,102] | |
| Reoviridae | Reovirus | σ1 | HSP70 and p23 | C′ trimerization of σ1 | Human | [103] |
| Rotavirus | NSP 3 | HSP90 | Dimerization of NSP3 | Human | [104] | |
| Rhabdoviridae | VSV | L protein | HSP90 | Protein stabilization and replication | Human | [48] |
| VSV | L protein and P protein | HSP60 | Synthesizes capped mRNAs and initiates transcription at the first gene (N) start site | Human | [105] | |
| Togaviridae | RUBV | p150 protein | HSP90 | Replication of the viral genome | Human | [106] |
| Tombusviridae | TBSV | Viral replicase | CNS1p cochaperone of HSP70 and HSP90 | Inhibited the assembly of the VRC and Viral RNA synthesis | Plant | [107] |
| TBSV | Viral replicase | HSP70 | Viral replication | Plant | [108] | |
| RCNMV | Viral replicase | HSP70 and HSP90 | Promoting virus replication | Plant | [109] | |
| RCNMV | p27 | HSP70 and HSP90 | Interaction with HSP70 or HSP90 | Plant | [109] | |
| Uncertain | RSVb | RdRp | HSP20 | Subcellular localization | Plant | [110] |
| RSVb | RdRp | HSP70 | Viral replication | Plant | [111] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Zhao, Y.; Wang, D.; Chen, Z. Mode of Action of Heat Shock Protein (HSP) Inhibitors against Viruses through Host HSP and Virus Interactions. Genes 2023, 14, 792. https://doi.org/10.3390/genes14040792
Wu S, Zhao Y, Wang D, Chen Z. Mode of Action of Heat Shock Protein (HSP) Inhibitors against Viruses through Host HSP and Virus Interactions. Genes. 2023; 14(4):792. https://doi.org/10.3390/genes14040792
Chicago/Turabian StyleWu, Shuang, Yongtian Zhao, Delu Wang, and Zhuo Chen. 2023. "Mode of Action of Heat Shock Protein (HSP) Inhibitors against Viruses through Host HSP and Virus Interactions" Genes 14, no. 4: 792. https://doi.org/10.3390/genes14040792
APA StyleWu, S., Zhao, Y., Wang, D., & Chen, Z. (2023). Mode of Action of Heat Shock Protein (HSP) Inhibitors against Viruses through Host HSP and Virus Interactions. Genes, 14(4), 792. https://doi.org/10.3390/genes14040792