Roles of miR-4442 in Colorectal Cancer: Predicting Early Recurrence and Regulating Epithelial-Mesenchymal Transition
Abstract
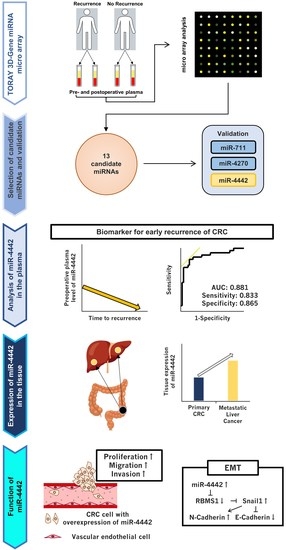
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Microarray Analysis
2.3. RNA Extraction from Plasma, Tissue, and Cell Line Sample
2.4. Quantitative Reverse Transcription–Polymerase Chain Reaction
2.5. Cell Lines and Cell Culture
2.6. Transfection of miRNA
2.7. Proliferation Assay
2.8. Migration and Invasion Assays
2.9. Prediction of Genes Targeted by miR-4442 In Silico
2.10. Luciferase Reporter Assay
2.11. Western Blot Analysis
2.12. Statistical Analysis
3. Results
3.1. Study Design
3.2. Selection of miRNA Candidates
3.3. Small- and Total-Scale Validation
3.4. Comparison of RFS, Clinicopathological Characteristics, and Plasma miR-4442 Levels
3.5. Subgroup Analysis of miR-4442 by Time to Recurrence and Diagnostic Ability of Plasma miR-4442 for Early Recurrence
3.6. Investigation into the Origin of miR-4442
3.7. Expression and Functions of miR-4442 in CRC Cell Line
3.8. Exploration of a Direct Target of miR-4442 and Evaluation of the Involvement of miR-4442 in the Regulation of EMT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Malakorn, S.; Ouchi, A.; Hu, C.Y.; Sandhu, L.; Dasari, A.; You, Y.N.; Kopetz, E.S.; Ellis, L.M.; Chang, G.J. Tumor Sidedness, Recurrence, and Survival After Curative Resection of Localized Colon Cancer. Clin. Color. Cancer 2021, 20, e53–e60. [Google Scholar] [CrossRef] [PubMed]
- Hansdotter, P.; Scherman, P.; Petersen, S.H.; Mikalonis, M.; Holmberg, E.; Rizell, M.; Naredi, P.; Syk, I. Patterns and resectability of colorectal cancer recurrences: Outcome study within the COLOFOL trial. BJS Open 2021, 5, zrab067. [Google Scholar] [CrossRef] [PubMed]
- Osterman, E.; Glimelius, B. Recurrence Risk After Up-to-Date Colon Cancer Staging, Surgery, and Pathology: Analysis of the Entire Swedish Population. Dis. Colon Rectum 2018, 61, 1016–1025. [Google Scholar] [CrossRef]
- Lan, Y.T.; Chang, S.C.; Yang, S.H.; Lin, C.C.; Wang, H.S.; Jiang, J.K.; Chen, W.S.; Lin, T.C.; Chiou, S.H.; Lin, J.K. Comparison of clinicopathological characteristics and prognosis between early and late recurrence after curative surgery for colorectal cancer. Am. J. Surg. 2014, 207, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.W.; Kim, C.H.; Lim, S.W.; Kim, H.R.; Kim, Y.J. Early recurrence in patients undergoing curative surgery for colorectal cancer: Is it a predictor for poor overall survival? Int. J. Color. Dis. 2013, 28, 1143–1149. [Google Scholar] [CrossRef]
- Furuke, H.; Arita, T.; Kuriu, Y.; Shimizu, H.; Kiuchi, J.; Yamamoto, Y.; Konishi, H.; Morimura, R.; Shiozaki, A.; Ikoma, H.; et al. The survival after recurrence of colorectal cancer: A retrospective study focused on time to recurrence after curative resection. Surg. Today 2021, 52, 239–250. [Google Scholar] [CrossRef]
- Sorensen, C.G.; Karlsson, W.K.; Pommergaard, H.C.; Burcharth, J.; Rosenberg, J. The diagnostic accuracy of carcinoembryonic antigen to detect colorectal cancer recurrence—A systematic review. Int. J. Surg. 2016, 25, 134–144. [Google Scholar] [CrossRef]
- Okamura, R.; Hasegawa, S.; Hida, K.; Hoshino, N.; Kawada, K.; Sugihara, K.; Sakai, Y.; The Japanese Study Group for Postoperative Follow-up of Colorectal Cancer. The role of periodic serum CA19-9 test in surveillance after colorectal cancer surgery. Int. J. Clin. Oncol. 2017, 22, 96–101. [Google Scholar] [CrossRef]
- Nicholson, B.D.; Shinkins, B.; Pathiraja, I.; Roberts, N.W.; James, T.J.; Mallett, S.; Perera, R.; Primrose, J.N.; Mant, D. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst. Rev. 2015, 2015, CD011134. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.Q.; Chen, C.; Peng, C.W.; Liu, S.P.; Li, Y. Carbohydrate antigen 242 highly consists with carbohydrate antigen 19-9 in diagnosis and prognosis of colorectal cancer: Study on 185 cases. Med. Oncol. 2012, 29, 1030–1036. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [Green Version]
- He, L.; He, X.; Lim, L.P.; de Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollis, M.; Nair, K.; Vyas, A.; Chaturvedi, L.S.; Gambhir, S.; Vyas, D. MicroRNAs potential utility in colon cancer: Early detection, prognosis, and chemosensitivity. World J. Gastroenterol. 2015, 21, 8284–8292. [Google Scholar] [CrossRef]
- Jung, G.; Hernandez-Illan, E.; Moreira, L.; Balaguer, F.; Goel, A. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 111–130. [Google Scholar] [CrossRef]
- To, K.K.; Tong, C.W.; Wu, M.; Cho, W.C. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J. Gastroenterol. 2018, 24, 2949–2973. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Hu, X.; Du, Y.; Du, J. The role of miRNAs in colorectal cancer progression and chemoradiotherapy. Biomed. Pharmacother. 2021, 134, 111099. [Google Scholar] [CrossRef] [PubMed]
- Balacescu, O.; Sur, D.; Cainap, C.; Visan, S.; Cruceriu, D.; Manzat-Saplacan, R.; Muresan, M.S.; Balacescu, L.; Lisencu, C.; Irimie, A. The Impact of miRNA in Colorectal Cancer Progression and Its Liver Metastases. Int. J. Mol. Sci. 2018, 19, 3711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Z.; Baker, K.; Redman, M.W.; Wang, L.; Adams, S.V.; Yu, M.; Dickinson, B.; Makar, K.; Ulrich, N.; Bohm, J.; et al. Dynamic plasma microRNAs are biomarkers for prognosis and early detection of recurrence in colorectal cancer. Br. J. Cancer 2017, 117, 1202–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyoshi, J.; Toden, S.; Yoshida, K.; Toiyama, Y.; Alberts, S.R.; Kusunoki, M.; Sinicrope, F.A.; Goel, A. MiR-139-5p as a novel serum biomarker for recurrence and metastasis in colorectal cancer. Sci. Rep. 2017, 7, 43393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brierley, J.; Gospodarowicz, M.; Wittekind, C. TNM Classification of Malignant Tumours, 8th ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017. [Google Scholar]
- Japanese Society for Cancer of the Colon and Rectum. Japanese Classification of Colorectal, Appendiceal, and Anal Carcinoma, 9th ed.; Kanehara & Co., Ltd.: Tokyo, Japan, 2018. [Google Scholar]
- Zou, A.; Liu, X.; Mai, Z.; Zhang, J.; Liu, Z.; Huang, Q.; Wu, A.; Zhou, C. LINC00472 Acts as a Tumor Suppressor in NSCLC through KLLN-Mediated p53-Signaling Pathway via MicroRNA-149-3p and MicroRNA-4270. Mol. Ther. Nucleic Acids 2019, 17, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, C.F.; Sun, L.B.; Li, Y.C. microRNA-4270-5p inhibits cancer cell proliferation and metastasis in hepatocellular carcinoma by targeting SATB2. Hum. Cell 2020, 33, 1155–1164. [Google Scholar] [CrossRef]
- Zhu, S.X.; Tong, X.Z.; Zhang, S. Expression of miR-711 and mechanism of proliferation and apoptosis in human gastric carcinoma. Oncol. Lett. 2017, 14, 4505–4510. [Google Scholar] [CrossRef] [Green Version]
- Xiao, W.S.; Li, D.F.; Tang, Y.P.; Chen, Y.Z.; Deng, W.B.; Chen, J.; Zhou, W.W.; Liao, A.J. Inhibition of epithelial-mesenchymal transition in gastric cancer cells by miR-711-mediated downregulation of CD44 expression. Oncol. Rep. 2018, 40, 2844–2853. [Google Scholar] [CrossRef]
- Das, D.K.; Persaud, L.; Sauane, M. MicroRNA-4719 and microRNA-6756-5p Correlate with Castration-Resistant Prostate Cancer Progression through Interleukin-24 Regulation. Noncoding RNA 2019, 5, 10. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; Li, Z.; Zhao, Y.; Song, A.; Shi, Y.; Hai, X.; Zhu, W. Breast cancer identification via modeling of peripherally circulating miRNAs. PeerJ 2018, 6, e4551. [Google Scholar] [CrossRef]
- Shen, D.; Zhao, H.; Zeng, P.; Song, J.; Yang, Y.; Gu, X.; Ji, Q.; Zhao, W. Circular RNA hsa_circ_0005556 Accelerates Gastric Cancer Progression by Sponging miR-4270 to Increase MMP19 Expression. J. Gastric Cancer 2020, 20, 300–312. [Google Scholar] [CrossRef]
- Li, L.; Gao, J.; Li, J.; Wang, J. MiR-711 regulates gastric cancer progression by targeting CD44. Cancer Biomark. 2022, 35, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Liao, A.; Tan, G.; Chen, L.; Zhou, W.; Hu, H. RASSF1A inhibits gastric cancer cell proliferation by miR-711- mediated downregulation of CDK4 expression. Oncotarget 2016, 7, 5842–5851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Zhao, Z.; Qi, Q.; Wang, J.; Kong, Y.; Feng, Z.; Chen, A.; Li, W.; Zhang, Q.; Wang, J.; et al. miR-6858 plays a key role in the process of melatonin inhibition of the malignant biological behavior of glioma. J. Clin. Neurosci. 2021, 87, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Hiraki, M.; Nishimura, J.; Takahashi, H.; Wu, X.; Takahashi, Y.; Miyo, M.; Nishida, N.; Uemura, M.; Hata, T.; Takemasa, I.; et al. Concurrent Targeting of KRAS and AKT by MiR-4689 Is a Novel Treatment Against Mutant KRAS Colorectal Cancer. Mol. Ther. Nucleic Acids 2015, 4, e231. [Google Scholar] [CrossRef]
- Mutlu, H.; Mutlu, S.; Bostanciklioglu, M. Profiling of Autophagy-Associated microRNAs in the Osteosarcoma Cell Line of U2OS. Anticancer Agents Med. Chem. 2021, 21, 1732–1737. [Google Scholar] [CrossRef]
- Yang, I.P.; Tsai, H.L.; Hou, M.F.; Chen, K.C.; Tsai, P.C.; Huang, S.W.; Chou, W.W.; Wang, J.Y.; Juo, S.H. MicroRNA-93 inhibits tumor growth and early relapse of human colorectal cancer by affecting genes involved in the cell cycle. Carcinogenesis 2012, 33, 1522–1530. [Google Scholar] [CrossRef] [Green Version]
- Tsai, H.L.; Yang, I.P.; Huang, C.W.; Ma, C.J.; Kuo, C.H.; Lu, C.Y.; Juo, S.H.; Wang, J.Y. Clinical significance of microRNA-148a in patients with early relapse of stage II stage and III colorectal cancer after curative resection. Transl. Res. 2013, 162, 258–268. [Google Scholar] [CrossRef]
- Hwang, C.C.; Chai, H.T.; Chen, H.W.; Tsai, H.L.; Lu, C.Y.; Yu, F.J.; Huang, M.Y.; Wang, J.Y. S100B protein expressions as an independent predictor of early relapse in UICC stages II and III colon cancer patients after curative resection. Ann. Surg. Oncol. 2011, 18, 139–145. [Google Scholar] [CrossRef]
- Zhou, J.; Zhan, S.; Tan, W.; Cheng, R.; Gong, H.; Zhu, Q. P300 binds to and acetylates MTA2 to promote colorectal cancer cells growth. Biochem. Biophys. Res. Commun. 2014, 444, 387–390. [Google Scholar] [CrossRef]
- Li, M.; Ma, Y.; Huang, P.; Du, A.; Yang, X.; Zhang, S.; Xing, C.; Liu, F.; Cao, J. Lentiviral DDX46 knockdown inhibits growth and induces apoptosis in human colorectal cancer cells. Gene 2015, 560, 237–244. [Google Scholar] [CrossRef]
- Chou, C.T.; Li, Y.J.; Chang, C.C.; Yang, C.N.; Li, P.S.; Jeng, Y.M.; Chen, S.T.; Kuo, M.L.; Lin, I.C.; Lin, B.R. Prognostic Significance of CDCP1 Expression in Colorectal Cancer and Effect of Its Inhibition on Invasion and Migration. Ann. Surg. Oncol. 2015, 22, 4335–4343. [Google Scholar] [CrossRef]
- Wang, H.; Huang, R.; Guo, W.; Qin, X.; Yang, Z.; Yuan, Z.; Wei, Y.; Mo, C.; Zeng, Z.; Luo, J.; et al. RNA-binding protein CELF1 enhances cell migration, invasion, and chemoresistance by targeting ETS2 in colorectal cancer. Clin. Sci. 2020, 134, 1973–1990. [Google Scholar] [CrossRef]
- Yu, J.; Navickas, A.; Asgharian, H.; Culbertson, B.; Fish, L.; Garcia, K.; Olegario, J.P.; Dermit, M.; Dodel, M.; Hanisch, B.; et al. RBMS1 Suppresses Colon Cancer Metastasis through Targeted Stabilization of Its mRNA Regulon. Cancer Discov. 2020, 10, 1410–1423. [Google Scholar] [CrossRef]
- Cojocneanu, R.; Braicu, C.; Raduly, L.; Jurj, A.; Zanoaga, O.; Magdo, L.; Irimie, A.; Muresan, M.S.; Ionescu, C.; Grigorescu, M.; et al. Plasma and Tissue Specific miRNA Expression Pattern and Functional Analysis Associated to Colorectal Cancer Patients. Cancers 2020, 12, 843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zen, K.; Zhang, C.Y. Circulating microRNAs: A novel class of biomarkers to diagnose and monitor human cancers. Med. Res. Rev. 2012, 32, 326–348. [Google Scholar] [CrossRef] [PubMed]
- AlSaieedi, A.; Salhi, A.; Tifratene, F.; Raies, A.B.; Hungler, A.; Uludag, M.; Van Neste, C.; Bajic, V.B.; Gojobori, T.; Essack, M. DES-Tcell is a knowledgebase for exploring immunology-related literature. Sci. Rep. 2021, 11, 14344. [Google Scholar] [CrossRef]
- Fisher, A.J.; Cipolla, E.; Varre, A.; Gu, H.; Mickler, E.A.; Vittal, R. Potential Mechanisms Underlying TGF-β-mediated Complement Activation in Lung Fibrosis. Cell. Mol. Med. Open Access 2017, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Dankert, J.T.; Wiesehofer, M.; Wach, S.; Czyrnik, E.D.; Wennemuth, G. Loss of RBMS1 as a regulatory target of miR-106b influences cell growth, gap closing and colony forming in prostate carcinoma. Sci. Rep. 2020, 10, 18022. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, P.; Bhavesh, N.S. Hinge like domain motion facilitates human RBMS1 protein binding to proto-oncogene c-myc promoter. Nucleic Acids Res. 2021, 49, 5943–5955. [Google Scholar] [CrossRef]
- Cha, J.H.; Wee, H.J.; Seo, J.H.; Ahn, B.J.; Park, J.H.; Yang, J.M.; Lee, S.W.; Lee, O.H.; Lee, H.J.; Gelman, I.H.; et al. Prompt meningeal reconstruction mediated by oxygen-sensitive AKAP12 scaffolding protein after central nervous system injury. Nat. Commun. 2014, 5, 4952. [Google Scholar] [CrossRef] [Green Version]
- Kaufhold, S.; Bonavida, B. Central role of Snail1 in the regulation of EMT and resistance in cancer: A target for therapeutic intervention. J. Exp. Clin. Cancer Res. 2014, 33, 62. [Google Scholar] [CrossRef] [PubMed]
- Young, P.E.; Womeldorph, C.M.; Johnson, E.K.; Maykel, J.A.; Brucher, B.; Stojadinovic, A.; Avital, I.; Nissan, A.; Steele, S.R. Early detection of colorectal cancer recurrence in patients undergoing surgery with curative intent: Current status and challenges. J. Cancer 2014, 5, 262–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Variable | Total (n = 108) | 3 yrs RFS (%) | p Value | p Value | HR | (95% CI) |
| Age (year) | ||||||
| ≥70 | 42 | 42.8 | 0.664 | |||
| <70 | 66 | 46.4 | ||||
| Sex | ||||||
| Male | 51 | 45.5 | 0.909 | |||
| Female | 57 | 44.6 | ||||
| Neoadjuvant chemotherapy | ||||||
| Present | 12 | 38.9 | 0.782 | |||
| Absent | 96 | 45.9 | ||||
| Adjuvant chemotherapy | ||||||
| Present | 45 | 33.7 | 0.182 | |||
| Absent | 63 | 52.2 | ||||
| Tumor location | ||||||
| Colon | 55 | 44.9 | 0.408 | |||
| Rectum | 53 | 43.3 | ||||
| Tumor type † | ||||||
| Differentiated type | 99 | 47.2 | 0.066 | |||
| Undifferentiated type | 9 | 22.2 | ||||
| Tumor size† (mm) | ||||||
| ≥40 | 54 | 30.4 | 0.016 | 0.271 | 1.404 | 0.771–2.630 |
| <40 | 54 | 59.5 | ||||
| Lymphatic invasion †‡ | ||||||
| Present | 61 | 38.3 | 0.043 | 0.788 | 1.087 | 0.583–1.982 |
| Absent | 47 | 53.8 | ||||
| Venous invasion †‡ | ||||||
| Present | 54 | 23.7 | <0.001 | 0.024 | 2.160 | 1.103–4.419 |
| Absent | 54 | 67.9 | ||||
| Tumor depth †§ | ||||||
| T3/4 | 78 | 34.9 | 0.002 | 0.867 | 1.077 | 0.461–2.675 |
| T1/2 | 30 | 65.5 | ||||
| Lymph node metastasis †§ | ||||||
| Positive | 56 | 24.5 | <0.001 | 0.026 | 1.931 | 1.080–3.600 |
| Negative | 52 | 66.7 | ||||
| CEA (ng/mL) | ||||||
| ≥5 | 54 | 31.4 | 0.006 | 0.528 | 1.205 | 0.679–2.178 |
| <5 | 54 | 57.7 | ||||
| CA19-9 (U/mL) | ||||||
| ≥37 | 12 | 8.3 | <0.001 | 0.097 | 1.927 | 0.883–3.934 |
| <37 | 96 | 50.1 | ||||
| Preoperative plasma level of miR-4442 | ||||||
| High (≥0.79) | 54 | 26.8 | <0.001 | 0.012 | 2.074 | 1.175–3.760 |
| Low (<0.79) | 54 | 63.7 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shibamoto, J.; Arita, T.; Konishi, H.; Kataoka, S.; Furuke, H.; Takaki, W.; Kiuchi, J.; Shimizu, H.; Yamamoto, Y.; Komatsu, S.; et al. Roles of miR-4442 in Colorectal Cancer: Predicting Early Recurrence and Regulating Epithelial-Mesenchymal Transition. Genes 2023, 14, 1414. https://doi.org/10.3390/genes14071414
Shibamoto J, Arita T, Konishi H, Kataoka S, Furuke H, Takaki W, Kiuchi J, Shimizu H, Yamamoto Y, Komatsu S, et al. Roles of miR-4442 in Colorectal Cancer: Predicting Early Recurrence and Regulating Epithelial-Mesenchymal Transition. Genes. 2023; 14(7):1414. https://doi.org/10.3390/genes14071414
Chicago/Turabian StyleShibamoto, Jun, Tomohiro Arita, Hirotaka Konishi, Satoshi Kataoka, Hirotaka Furuke, Wataru Takaki, Jun Kiuchi, Hiroki Shimizu, Yusuke Yamamoto, Shuhei Komatsu, and et al. 2023. "Roles of miR-4442 in Colorectal Cancer: Predicting Early Recurrence and Regulating Epithelial-Mesenchymal Transition" Genes 14, no. 7: 1414. https://doi.org/10.3390/genes14071414
APA StyleShibamoto, J., Arita, T., Konishi, H., Kataoka, S., Furuke, H., Takaki, W., Kiuchi, J., Shimizu, H., Yamamoto, Y., Komatsu, S., Shiozaki, A., Kuriu, Y., & Otsuji, E. (2023). Roles of miR-4442 in Colorectal Cancer: Predicting Early Recurrence and Regulating Epithelial-Mesenchymal Transition. Genes, 14(7), 1414. https://doi.org/10.3390/genes14071414