Transmission Dynamics of HIV-1 Drug Resistance among Treatment-Naïve Individuals in Greece: The Added Value of Molecular Epidemiology to Public Health
Abstract
:1. Introduction
2. Materials and Methods
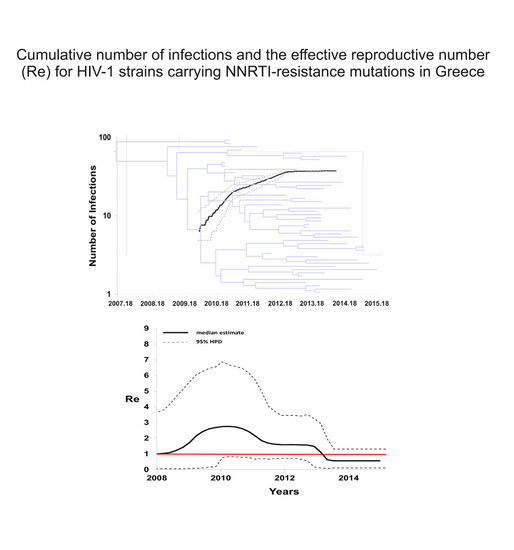
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cozzi-Lepri, A.; Noguera-Julian, M.; Di Giallonardo, F.; Schuurman, R.; Daumer, M.; Aitken, S.; Ceccherini-Silberstein, F.; D’Arminio Monforte, A.; Geretti, A.M.; Booth, C.L.; et al. Low-frequency drug-resistant HIV-1 and risk of virological failure to first-line NNRTI-based art: A multicohort european case-control study using centralized ultrasensitive 454 pyrosequencing. J. Antimicrob. Chemother. 2015, 70, 930–940. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Paredes, R.; Ribaudo, H.J.; Svarovskaia, E.S.; Metzner, K.J.; Kozal, M.J.; Hullsiek, K.H.; Balduin, M.; Jakobsen, M.R.; Geretti, A.M.; et al. Low-frequency HIV-1 drug resistance mutations and risk of nnrti-based antiretroviral treatment failure: A systematic review and pooled analysis. JAMA 2011, 305, 1327–1335. [Google Scholar] [CrossRef] [PubMed]
- Wittkop, L.; Gunthard, H.F.; de Wolf, F.; Dunn, D.; Cozzi-Lepri, A.; de Luca, A.; Kucherer, C.; Obel, N.; von Wyl, V.; Masquelier, B.; et al. Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (eurocoord-chain joint project): A european multicohort study. Lancet Infect. Dis. 2011, 11, 363–371. [Google Scholar] [CrossRef]
- Wensing, A.M.; Calvez, V.; Gunthard, H.F.; Johnson, V.A.; Paredes, R.; Pillay, D.; Shafer, R.W.; Richman, D.D. 2017 update of the drug resistance mutations in HIV-1. Top. Antivir. Med. 2017, 24, 132–133. [Google Scholar] [PubMed]
- Rhee, S.Y.; Blanco, J.L.; Jordan, M.R.; Taylor, J.; Lemey, P.; Varghese, V.; Hamers, R.L.; Bertagnolio, S.; de Wit, T.F.; Aghokeng, A.F.; et al. Correction: Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: An individual-patient- and sequence-level meta-analysis. PLoS Med. 2015, 12, e1001845. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.Y.; Blanco, J.L.; Jordan, M.R.; Taylor, J.; Lemey, P.; Varghese, V.; Hamers, R.L.; Bertagnolio, S.; Rinke de Wit, T.F.; Aghokeng, A.F.; et al. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: An individual-patient- and sequence-level meta-analysis. PLoS Med. 2015, 12, e1001810. [Google Scholar] [CrossRef] [Green Version]
- Balode, D.; Westman, M.; Kolupajeva, T.; Rozentale, B.; Albert, J. Low prevalence of transmitted drug resistance among newly diagnosed HIV-1 patients in Latvia. J. Med. Virol. 2010, 82, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- SPREAD programme. Transmission of drug-resistant HIV-1 in Europe remains limited to single classes. AIDS 2008, 22, 625–635. [Google Scholar] [CrossRef]
- Hofstra, L.M.; Sauvageot, N.; Albert, J.; Alexiev, I.; Garcia, F.; Struck, D.; Van de Vijver, D.A.; Asjo, B.; Beshkov, D.; Coughlan, S.; et al. Transmission of HIV drug resistance and the predicted effect on current first-line regimens in Europe. Clin. Infect. Dis. 2016, 62, 655–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wensing, A.M.; van de Vijver, D.A.; Angarano, G.; Asjo, B.; Balotta, C.; Boeri, E.; Camacho, R.; Chaix, M.L.; Costagliola, D.; De Luca, A.; et al. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: Implications for clinical management. J. Infect. Dis. 2005, 192, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Castor, D.; Low, A.; Evering, T.; Karmon, S.; Davis, B.; Figueroa, A.; LaMar, M.; Garmon, D.; Mehandru, S.; Markowitz, M. Transmitted drug resistance and phylogenetic relationships among acute and early HIV-1-infected individuals in New York city. J. Acquir. Immune Defic. Syndr. 2012, 61, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Boden, D.; Hurley, A.; Zhang, L.; Cao, Y.; Guo, Y.; Jones, E.; Tsay, J.; Ip, J.; Farthing, C.; Limoli, K.; et al. HIV-1 drug resistance in newly infected individuals. JAMA 1999, 282, 1135–1141. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.G.; Roger, M.; Moisi, D.D.; Oliveira, M.; Hardy, I.; Turgel, R.; Charest, H.; Routy, J.P.; Wainberg, M.A. Transmission networks of drug resistance acquired in primary/early stage HIV infection. AIDS 2008, 22, 2509–2515. [Google Scholar] [CrossRef] [PubMed]
- Toni, T.A.; Asahchop, E.L.; Moisi, D.; Ntemgwa, M.; Oliveira, M.; Masquelier, B.; Brenner, B.G.; Wainberg, M.A. Detection of human immunodeficiency virus (HIV) type 1 M184V and K103N minority variants in patients with primary HIV infection. Antimicrob. Agents Chemother. 2009, 53, 1670–1672. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.G.; Roger, M.; Routy, J.P.; Moisi, D.; Ntemgwa, M.; Matte, C.; Baril, J.G.; Thomas, R.; Rouleau, D.; Bruneau, J.; et al. High rates of forward transmission events after acute/early HIV-1 infection. J. Infect. Dis. 2007, 195, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Paraskevis, D.; Kostaki, E.; Magiorkinis, G.; Gargalianos, P.; Xylomenos, G.; Magiorkinis, E.; Lazanas, M.; Chini, M.; Nikolopoulos, G.; Skoutelis, A.; et al. Prevalence of drug resistance among HIV-1 treatment-naïve patients in Greece during 2003–2015: Transmitted drug resistance is due to onward transmissions. Infect. Genet. Evol. 2017, 54, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Paraskevis, D.; Nikolopoulos, G.; Tsiara, C.; Paraskeva, D.; Antoniadou, A.; Lazanas, M.; Gargalianos, P.; Psychogiou, M.; Malliori, M.; Kremastinou, J.; et al. HIV-1 outbreak among injecting drug users in Greece, 2011: A preliminary report. Euro Surveill. 2011, 16, 19962. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Heled, J.; Kuhnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [PubMed]
- Stadler, T.; Kuhnert, D.; Bonhoeffer, S.; Drummond, A.J. Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV). Proc. Natl. Acad. Sci. USA 2013, 110, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Paraskevis, D.; Paraschiv, S.; Sypsa, V.; Nikolopoulos, G.; Tsiara, C.; Magiorkinis, G.; Psichogiou, M.; Flampouris, A.; Mardarescu, M.; Niculescu, I.; et al. Enhanced HIV-1 surveillance using molecular epidemiology to study and monitor HIV-1 outbreaks among intravenous drug users (IDUS) in Athens and Bucharest. Infect. Genet. Evol. 2015, 35, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.L.; Kouyos, R.D.; Boni, J.; Yerly, S.; Klimkait, T.; Aubert, V.; Scherrer, A.U.; Shilaih, M.; Hinkley, T.; Petropoulos, C.; et al. Persistence of transmitted HIV-1 drug resistance mutations associated with fitness costs and viral genetic backgrounds. PLoS Pathog. 2015, 11, e1004722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European AIDS Clinical Society. Guidelines, version 8.2; European AIDS Clinical Society: Brussels, Belgium, 2017. [Google Scholar]
- Antoniadou, Z.A.; Kousiappa, I.; Skoura, L.; Pilalas, D.; Metallidis, S.; Nicolaidis, P.; Malisiovas, N.; Kostrikis, L.G. Short communication: Molecular epidemiology of HIV type 1 infection in northern Greece (2009–2010): Evidence of a transmission cluster of HIV type 1 subtype a1 drug-resistant strains among men who have sex with men. AIDS Res. Hum. Retrovir. 2014, 30, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Drescher, S.M.; von Wyl, V.; Yang, W.L.; Boni, J.; Yerly, S.; Shah, C.; Aubert, V.; Klimkait, T.; Taffe, P.; Furrer, H.; et al. Treatment-naive individuals are the major source of transmitted HIV-1 drug resistance in men who have sex with men in the swiss HIV cohort study. Clin. Infect. Dis. 2014, 58, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Mbisa, J.L.; Fearnhill, E.; Dunn, D.T.; Pillay, D.; Asboe, D.; Cane, P.A. Evidence of self-sustaining drug resistant HIV-1 lineages among untreated patients in the United Kingdom. Clin. Infect. Dis. 2015, 61, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Mourad, R.; Chevennet, F.; Dunn, D.T.; Fearnhill, E.; Delpech, V.; Asboe, D.; Gascuel, O.; Hue, S. A phylotype-based analysis highlights the role of drug-naive HIV-positive individuals in the transmission of antiretroviral resistance in the UK. AIDS 2015, 29, 1917–1925. [Google Scholar] [CrossRef] [PubMed]
- Theys, K.; Van Laethem, K.; Gomes, P.; Baele, G.; Pineda-Pena, A.C.; Vandamme, A.M.; Camacho, R.J.; Abecasis, A.B. Sub-epidemics explain localized high prevalence of reduced susceptibility to rilpivirine in treatment-naive HIV-1-infected patients: Subtype and geographic compartmentalization of baseline resistance mutations. AIDS Res. Hum. Retrovir. 2016, 32, 427–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Local Transmission Network | Number of Individuals | tMRCA 1 (Median; 95% HPD 2) | Re 3 (Maximum Value of Median) | CV 4 (Median; 95% HPD) |
|---|---|---|---|---|
| K103N | 48 | 2007 (2004–2009) | 2.8 | 0.25 (0.00002–0.68) |
| E138A_1 | 22 | 1995 (1991–1999) | 2.1 | 1.10 (0.57–1.72) |
| E138A_2 | 38 | 1996 (1989–2000) | 1.8 | 0.73 (0.47–1.04) |
| E138A_3 | 50 | 1997 (1991–2001) | 2.0 | 0.84 (0.48–1.24) |
| E138A_4 | 38 | 2004 (2000–2007) | 2.5 | 0.58 (0.20–0.95) |
| Local Transmission Networks | ||||||
|---|---|---|---|---|---|---|
| K103N (N, %) | E138A_1 (N, %) | E138A_2 (N, %) | E138A_3 (N, %) | E138A_4 (N, %) | p-value | |
| Number of individuals | 48 (24.5) | 22 (11.2) | 38 (19.4) | 50 (25.5) | 38 (19.4) | |
| Gender | 0.159 | |||||
| Male | 48 (100.0) | 21 (95.5) | 36 (94.7) | 50 (100.0) | 37 (97.4) | |
| Female | 0 (0.0) | 1 (4.5) | 2 (5.3) | 0 (0.0) | 1 (2.6) | |
| Risk group | 0.046 | |||||
| MSM 1 | 35 (72.9) | 13 (59.1) | 24 (63.2) | 39 (78.0) | 32 (84.2) | |
| PWID 2 | 1 (2.1) | 0 (0.0) | 1 (2.6) | 3 (6.0) | 1 (2.6) | |
| MSW 3 | 1 (2.1) | 5 (22.7) | 5 (13.2) | 3 (6.0) | 0 (0.0) | |
| Unknown | 11 (22.9) | 4 (18.2) | 8 (21.0) | 5 (10.0) | 5 (13.2) | |
| Nationality | 0.020 | |||||
| Hellenic | 21 (43.8) | 12 (54.6) | 27 (71.1) | 37 (74.0) | 21 (55.3) | |
| Non Hellenic | 2 (4.2) | 0 (0.0) | 3 (7.9) | 1 (2.0) | 1 (2.6) | |
| Unknown | 25 (52.0) | 10 (45.4) | 8 (21.0) | 12 (24.0) | 16 (42.1) | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paraskevis, D.; Kostaki, E.; Gargalianos, P.; Xylomenos, G.; Lazanas, M.; Chini, M.; Skoutelis, A.; Papastamopoulos, V.; Paraskeva, D.; Antoniadou, A.; et al. Transmission Dynamics of HIV-1 Drug Resistance among Treatment-Naïve Individuals in Greece: The Added Value of Molecular Epidemiology to Public Health. Genes 2017, 8, 322. https://doi.org/10.3390/genes8110322
Paraskevis D, Kostaki E, Gargalianos P, Xylomenos G, Lazanas M, Chini M, Skoutelis A, Papastamopoulos V, Paraskeva D, Antoniadou A, et al. Transmission Dynamics of HIV-1 Drug Resistance among Treatment-Naïve Individuals in Greece: The Added Value of Molecular Epidemiology to Public Health. Genes. 2017; 8(11):322. https://doi.org/10.3390/genes8110322
Chicago/Turabian StyleParaskevis, Dimitrios, Evangelia Kostaki, Panagiotis Gargalianos, Georgios Xylomenos, Marios Lazanas, Maria Chini, Athanasios Skoutelis, Vasileios Papastamopoulos, Dimitra Paraskeva, Anastasia Antoniadou, and et al. 2017. "Transmission Dynamics of HIV-1 Drug Resistance among Treatment-Naïve Individuals in Greece: The Added Value of Molecular Epidemiology to Public Health" Genes 8, no. 11: 322. https://doi.org/10.3390/genes8110322
APA StyleParaskevis, D., Kostaki, E., Gargalianos, P., Xylomenos, G., Lazanas, M., Chini, M., Skoutelis, A., Papastamopoulos, V., Paraskeva, D., Antoniadou, A., Papadopoulos, A., Psichogiou, M., Daikos, G. L., Chrysos, G., Paparizos, V., Kourkounti, S., Sambatakou, H., Sipsas, N. V., Lada, M., ... Hatzakis, A. (2017). Transmission Dynamics of HIV-1 Drug Resistance among Treatment-Naïve Individuals in Greece: The Added Value of Molecular Epidemiology to Public Health. Genes, 8(11), 322. https://doi.org/10.3390/genes8110322