The Emerging Role of Epitranscriptomics in Cancer: Focus on Urological Tumors
Abstract
:1. RNA Modifications in Brief: From Epigenetics to Epitranscriptomics
2. m6A Modification in Non-Urological Malignancies: Literature Review
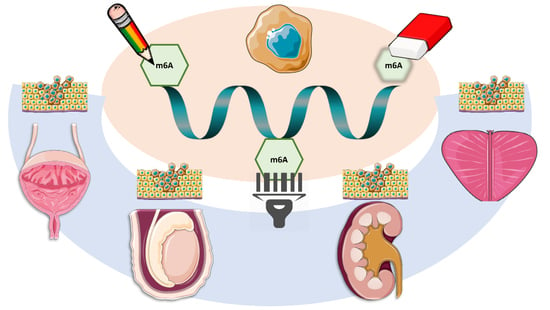
3. m6A Modifications in Urological Tumors: Analysis of The Cancer Genome Atlas Database
3.1. Prostate Cancer
3.2. Testicular Cancer
3.3. Kidney Cancer
3.4. Bladder Cancer
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Bheda, P.; Schneider, R. Epigenetics reloaded: The single-cell revolution. Trends Cell Biol. 2014, 24, 712–723. [Google Scholar] [CrossRef] [PubMed]
- He, C. Grand challenge commentary: RNA epigenetics? Nat. Chem. Biol. 2010, 6, 863–865. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J. The RNA modification N(6)-methyladenosine and its implications in human disease. Genom. Proteom. Bioinform. 2017, 15, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Park, E.; Lin, L.; Xing, Y. A panoramic view of RNA modifications: Exploring new frontiers. Genome Biol. 2018, 19, 11. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; De Crecy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef] [PubMed]
- Davalos, V.; Blanco, S.; Esteller, M. SnapShot: Messenger RNA modifications. Cell 2018, 174, 498–498.e1. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jia, G. Methylation modifications in eukaryotic messenger RNA. J. Genet. Genom. 2014, 41, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.; Zander, S.; Gutschner, T. The dark side of the epitranscriptome: Chemical modifications in long non-coding RNAs. Int. J. Mol. Sci. 2017, 18, 2387. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Jaffrey, S.R.; Pan, T.; Rechavi, G.; Suzuki, T. RNA modifications: What have we learned and where are we headed? Nat. Rev. Genet. 2016, 17, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Yi, C.; Peng, J. Epitranscriptomics: Toward a better understanding of RNA modifications. Genom. Proteom. Bioinform. 2017, 15, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Pan, T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem. Sci. 2013, 38, 204–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Pan, T. N6-methyladenosine-encoded epitranscriptomics. Nat. Struct. Mol. Biol. 2016, 23, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maity, A.; Das, B. N6-methyladenosine modification in mRNA: Machinery, function and implications for health and diseases. FEBS J. 2016, 283, 1607–1630. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, X.; Yi, C. Epitranscriptome sequencing technologies: Decoding RNA modifications. Nat. Methods 2016, 14, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Imanishi, M.; Tsuji, S.; Suda, A.; Futaki, S. Detection of N(6)-methyladenosine based on the methyl-sensitivity of MazF RNA endonuclease. Chem. Commun. (Camb.) 2017, 53, 12930–12933. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Kapoor, U.; Jantsch, M.F. Understanding RNA modifications: The promises and technological bottlenecks of the ‘epitranscriptome’. Open Biol. 2017, 7, 170077. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.P.; Pickering, B.F.; Jaffrey, S.R. Reading m(6)A in the transcriptome: M(6)A-binding proteins. Trends Cell Biol. 2018, 28, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, A.; Somasundaram, K. mRNA traffic control reviewed: N6-methyladenosine (m(6)A) takes the driver’s seat. Bioessays 2018, 40. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Li, L.; Huang, Y.; Ma, J.; Min, J. Readers, writers and erasers of N(6)-methylated adenosine modification. Curr. Opin. Struct. Biol. 2017, 47, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yin, P. Structural Insights into N(6)-methyladenosine (m(6)A) modification in the transcriptome. Genom. Proteom. Bioinform. 2018, 16, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Qiao, J.; Wang, G.; Lan, Y.; Li, G.; Guo, X.; Xi, J.; Ye, D.; Zhu, S.; Chen, W.; et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018, 46, 3906–3920. [Google Scholar] [CrossRef] [PubMed]
- Bertero, A.; Brown, S.; Madrigal, P.; Osnato, A.; Ortmann, D.; Yiangou, L.; Kadiwala, J.; Hubner, N.C.; De Los Mozos, I.R.; Sadee, C.; et al. The SMAD2/3 interactome reveals that TGFbeta controls m(6)A mRNA methylation in pluripotency. Nature 2018, 555, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.L.; Wang, X.; An, R.; Cassin, J.; Vissers, C.; Liu, Y.; Liu, Y.; Xu, T.; Wang, X.; Wong, S.Z.H.; et al. Epitranscriptomic m(6)A regulation of axon regeneration in the adult mammalian nervous system. Neuron 2018, 97, 313–325.e6. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Cao, G.; Zhang, T.; Sefik, E.; Amezcua Vesely, M.C.; Broughton, J.P.; Zhu, S.; Li, H.; Li, B.; Chen, L.; et al. m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res. 2018, 28, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.; Chen, A. The emerging role of mRNA methylation in normal and pathological behavior. Genes Brain Behav. 2017, 17, e12428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.; Zhang, Z.; Yu, L.; Li, Y.; Liang, M.; Zhou, L. FTO reduces mitochondria and promotes hepatic fat accumulation through RNA demethylation. J. Cell. Biochem. 2018, 119, 5676–5685. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Zou, Q.; Ding, J.; Ling, M.; Wang, W.; Li, H.; Huang, B. Increased N6-Methyladenosine causes infertility is associated with FTO expression. J. Cell. Physiol. 2018, 233, 7055–7066. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichinchi, G.; Gao, S.; Saletore, Y.; Gonzalez, G.M.; Bansal, V.; Wang, Y.; Mason, C.E.; Rana, T.M. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016, 1, 16011. [Google Scholar] [CrossRef] [PubMed]
- Fustin, J.M.; Doi, M.; Yamaguchi, Y.; Hida, H.; Nishimura, S.; Yoshida, M.; Isagawa, T.; Morioka, M.S.; Kakeya, H.; Manabe, I.; et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013, 155, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Shih, J.; Hollern, D.P.; Wang, L.; Bowlby, R.; Tickoo, S.K.; Thorsson, V.; Mungall, A.J.; Newton, Y.; Hegde, A.M.; et al. Integrated molecular characterization of testicular germ cell tumors. Cell Rep. 2018, 23, 3392–3406. [Google Scholar] [CrossRef] [PubMed]
- Tusup, M.; Kundig, T.; Pascolo, S. Epitranscriptomics of cancer. World J. Clin. Oncol. 2018, 9, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chai, P.; Jia, R.; Jia, R. Novel insights on m(6)A RNA methylation in tumorigenesis: A double-edged sword. Mol. Cancer 2018, 17, 101. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Pandolfi, P.P. The epitranscriptome of noncoding RNAs in cancer. Cancer Discov. 2017, 7, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.X.; Li, L.M.; Sun, H.L.; Liu, S.M. Link between m6A modification and cancers. Front. Bioeng. Biotechnol. 2018, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Sun, C.; Li, J.; Zhang, E.; Ma, Z.; Xu, W.; Li, H.; Qiu, M.; Xu, Y.; Xia, W.; et al. Roles of RNA methylation by means of N(6)-methyladenosine (m(6)A) in human cancers. Cancer Lett. 2017, 408, 112–120. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Li, J.; Wang, X.; Ying, Y.; Xie, H.; Yan, H.; Zheng, X.; Xie, L. The dual role of N6-methyladenosine modification of RNAs is involved in human cancers. J. Cell Mol. Med. 2018, 22, 4630–4639. [Google Scholar] [CrossRef] [PubMed]
- Lian, H.; Wang, Q.H.; Zhu, C.B.; Ma, J.; Jin, W.L. Deciphering the epitranscriptome in cancer. Trends Cancer 2018, 4, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Liu, K.; Tempel, W.; Demetriades, M.; Aik, W.; Schofield, C.J.; Min, J. Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J. Biol. Chem. 2014, 289, 17299–17311. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Chen, Q.; Wang, L.; Ke, D.; Yuan, Z. Association between FTO gene polymorphism and cancer risk: Evidence from 16,277 cases and 31,153 controls. Tumour Biol. 2012, 33, 1237–1243. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Frye, M. Role of RNA methyltransferases in tissue renewal and pathology. Curr. Opin. Cell Biol. 2014, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Iles, M.M.; Law, M.H.; Stacey, S.N.; Han, J.; Fang, S.; Pfeiffer, R.; Harland, M.; Macgregor, S.; Taylor, J.C.; Aben, K.K.; et al. A variant in FTO shows association with melanoma risk not due to BMI. Nat. Genet. 2013, 45, 428–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Closas, M.; Couch, F.J.; Lindstrom, S.; Michailidou, K.; Schmidt, M.K.; Brook, M.N.; Orr, N.; Rhie, S.K.; Riboli, E.; Feigelson, H.S.; et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat. Genet. 2013, 45, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.; Dang, Y.; Chen, G.; Mo, Z. Overexpression of the fat mass and obesity associated gene (FTO) in breast cancer and its clinical implications. Int. J. Clin. Exp. Pathol. 2015, 8, 13405–13410. [Google Scholar] [PubMed]
- Singh, B.; Kinne, H.E.; Milligan, R.D.; Washburn, L.J.; Olsen, M.; Lucci, A. Important role of FTO in the survival of rare panresistant triple-negative inflammatory breast cancer cells facing a severe metabolic challenge. PLoS ONE 2016, 11, e0159072. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhi, W.I.; Lu, H.; Samanta, D.; Chen, I.; Gabrielson, E.; Semenza, G.L. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget 2016, 7, 64527–64542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Samanta, D.; Lu, H.; Bullen, J.W.; Zhang, H.; Chen, I.; He, X.; Semenza, G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E2047–E2056. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Wang, X.; Cao, C.; Gao, Y.; Zhang, S.; Yang, Z.; Liu, Y.; Zhang, X.; Zhang, W.; Ye, L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018, 415, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wei, L.; Law, C.T.; Tsang, F.H.; Shen, J.; Cheng, C.L.; Tsang, L.H.; Ho, D.W.; Chiu, D.K.; Lee, J.M.; et al. RNA N6-methyladenosine methyltransferase METTL3 promotes liver cancer progression through YTHDF2 dependent post-transcriptional silencing of SOCS2. Hepatology 2017, 67, 2254–2270. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, Y.; Mao, Q.; Jiang, X.; Jiang, W.; Chen, J.; Xu, W.; Zhong, L.; Sun, X. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018, 21, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.Z.; Yang, F.; Zhou, C.C.; Liu, F.; Yuan, J.H.; Wang, F.; Wang, T.T.; Xu, Q.G.; Zhou, W.P.; Sun, S.H. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6)-methyladenosine-dependent primary microRNA processing. Hepatology 2017, 65, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Taketo, K.; Konno, M.; Asai, A.; Koseki, J.; Toratani, M.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H.; Ogawa, K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 2018, 52, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Sun, Y.; Xu, X.; Wang, D.; He, J.; Zhou, H.; Lu, Y.; Zeng, J.; Du, F.; Gong, A.; et al. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle 2017, 16, 2259–2271. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.J.; Shim, H.E.; Han, M.E.; Kim, H.J.; Kim, K.S.; Baek, S.; Choi, K.U.; Hur, G.Y.; Oh, S.O. WTAP regulates migration and invasion of cholangiocarcinoma cells. J. Gastroenterol. 2013, 48, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, Y.; Konno, M.; Asai, A.; Koseki, J.; Kawamoto, K.; Miyoshi, N.; Takahashi, H.; Nishida, N.; Haraguchi, N.; Sakai, D.; et al. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget 2018, 9, 7476–7486. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, A.; Tanikawa, K.; Tsunetomi, M.; Takai, K.; Ikeda, H.; Konno, J.; Torigoe, T.; Maeda, H.; Kutomi, G.; Okita, K.; et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1alpha mRNA is translated. Cancer Lett. 2016, 376, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Porcellini, E.; Laprovitera, N.; Riefolo, M.; Ravaioli, M.; Garajova, I.; Ferracin, M. Epigenetic and epitranscriptomic changes in colorectal cancer: Diagnostic, prognostic, and treatment implications. Cancer Lett. 2018, 419, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Shao, W.; Jiang, Y.; Wang, X.; Liu, Y.; Liu, X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol. Rep. 2017, 38, 2285–2292. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Z.; Kong, B.; Song, C.; Cong, J.; Hou, J.; Wang, S. Reduced m(6)A mRNA methylation is correlated with the progression of human cervical cancer. Oncotarget 2017, 8, 98918–98930. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Bai, Z.L.; Xia, D.; Zhao, Z.J.; Zhao, R.; Wang, Y.Y.; Zhe, H. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol. Carcinog. 2018, 57, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Eckert, M.A.; Harada, B.T.; Liu, S.M.; Lu, Z.; Yu, K.; Tienda, S.M.; Chryplewicz, A.; Zhu, A.C.; Yang, Y.; et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018, 20, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.G.; et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017, 18, 2622–2634. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bogler, O.; et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 2017, 31, 591–606.e6. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, A.; Patil, V.; Arora, A.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Somasundaram, K. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 2018, 37, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Zhang, Y.; Mao, Y.; Mou, J.; Zhao, J.; Xue, Q.; Wang, D.; Huang, J.; Gao, S.; Gao, Y. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem. Biophys. Res. Commun. 2017, 482, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Kwok, C.T.; Marshall, A.D.; Rasko, J.E.; Wong, J.J. Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. J. Hematol. Oncol. 2017, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Weng, H.; Huang, H.; Wu, H.; Qin, X.; Zhao, B.S.; Dong, L.; Shi, H.; Skibbe, J.; Shen, C.; Hu, C.; et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 2018, 22, 191–205.e9. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, I.; Tzelepis, K.; Pandolfini, L.; Shi, J.; Millan-Zambrano, G.; Robson, S.C.; Aspris, D.; Migliori, V.; Bannister, A.J.; Han, N.; et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 2017, 552, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; MacKay, M.; et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Bansal, H.; Yihua, Q.; Iyer, S.P.; Ganapathy, S.; Proia, D.A.; Penalva, L.O.; Uren, P.J.; Suresh, U.; Carew, J.S.; Karnad, A.B.; et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia 2014, 28, 1171–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Z.; Morris, S.W.; Valentine, V.; Li, M.; Herbrick, J.A.; Cui, X.; Bouman, D.; Li, Y.; Mehta, P.K.; Nizetic, D.; et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 2001, 28, 220–221. [Google Scholar] [CrossRef] [PubMed]
- Casalegno-Garduno, R.; Schmitt, A.; Wang, X.; Xu, X.; Schmitt, M. Wilms’ tumor 1 as a novel target for immunotherapy of leukemia. Transplant. Proc. 2010, 42, 3309–3311. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Cairns, R.A.; Mak, T.W. Oncogenic isocitrate dehydrogenase mutations: Mechanisms, models, and clinical opportunities. Cancer Discov. 2013, 3, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Elkashef, S.M.; Lin, A.P.; Myers, J.; Sill, H.; Jiang, D.; Dahia, P.L.M.; Aguiar, R.C.T. IDH Mutation, competitive inhibition of FTO, and RNA methylation. Cancer Cell 2017, 31, 619–620. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Center, M.M.; Jemal, A.; Lortet-Tieulent, J.; Ward, E.; Ferlay, J.; Brawley, O.; Bray, F. International variation in prostate cancer incidence and mortality rates. Eur. Urol. 2012, 61, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Roobol, M.J.; Carlsson, S.V. Risk stratification in prostate cancer screening. Nat. Rev. Urol. 2013, 10, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.M.; Abate-Shen, C. Molecular genetics of prostate cancer: New prospects for old challenges. Genes Dev. 2010, 24, 1967–2000. [Google Scholar] [CrossRef] [PubMed]
- Ceder, Y.; Bjartell, A.; Culig, Z.; Rubin, M.A.; Tomlins, S.; Visakorpi, T. The molecular evolution of castration-resistant prostate cancer. Eur. Urol. Focus 2016, 2, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Sardana, G.; Diamandis, E.P. Biomarkers for the diagnosis of new and recurrent prostate cancer. Biomark. Med. 2012, 6, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Chiam, K.; Ricciardelli, C.; Bianco-Miotto, T. Epigenetic biomarkers in prostate cancer: Current and future uses. Cancer Lett. 2014, 342, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Meng, S.; Xu, M.; Wang, S.; He, L.; Xu, X.; Wang, X.; Xie, L. Downregulation of N6-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N6-methyladenosine levels. Oncotarget 2018, 9, 3752–3764. [Google Scholar] [PubMed]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO classification of tumours of the urinary system and male genital organs-part A: Renal, penile, and testicular tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Trabert, B.; Chen, J.; Devesa, S.S.; Bray, F.; McGlynn, K.A. International patterns and trends in testicular cancer incidence, overall and by histologic subtype, 1973–2007. Andrology 2015, 3, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Beyer, J.; Albers, P.; Altena, R.; Aparicio, J.; Bokemeyer, C.; Busch, J.; Cathomas, R.; Cavallin-Stahl, E.; Clarke, N.W.; Classen, J.; et al. Maintaining success, reducing treatment burden, focusing on survivorship: Highlights from the third European consensus conference on diagnosis and treatment of germ-cell cancer. Ann. Oncol. 2013, 24, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.L.; Lobo, J.; Jeronimo, C.; Henrique, R. The epigenetics of testicular germ cell tumors: Looking for novel disease biomarkers. Epigenomics 2017, 9, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Lobo, J.; Costa, A.L.; Vilela-Salgueiro, B.; Rodrigues, A.; Guimaraes, R.; Cantante, M.; Lopes, P.; Antunes, L.; Jeronimo, C.; Henrique, R. Testicular germ cell tumors: Revisiting a series in light of the new WHO classification and AJCC staging systems, focusing on challenges for pathologists. Hum. Pathol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Henrique, R.; Jeronimo, C. Testicular germ cell tumors go epigenetics: Will miR-371a-3p replace classical serum biomarkers? Eur. Urol. 2017, 71, 221–222. [Google Scholar] [CrossRef] [PubMed]
- Aoun, F.; Kourie, H.R.; Albisinni, S.; Roumeguere, T. Will testicular germ cell tumors remain untargetable? Target Oncol. 2016, 11, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Boccellino, M.; Vanacore, D.; Zappavigna, S.; Cavaliere, C.; Rossetti, S.; D’Aniello, C.; Chieffi, P.; Amler, E.; Buonerba, C.; Di Lorenzo, G.; et al. Testicular cancer from diagnosis to epigenetic factors. Oncotarget 2017, 8, 104654–104663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, M.J.; Huddart, R.A.; Coleman, N. The present and future of serum diagnostic tests for testicular germ cell tumours. Nat. Rev. Urol. 2016, 13, 715–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Der Zwan, Y.G.; Stoop, H.; Rossello, F.; White, S.J.; Looijenga, L.H. Role of epigenetics in the etiology of germ cell cancer. Int. J. Dev. Biol. 2013, 57, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, P.M.; Read, G. International Germ Cell Consensus Classification: A prognostic factor-based staging system for metastatic germ cell cancers. International Germ Cell Cancer Collaborative Group. J. Clin. Oncol. 1997, 15, 594–603. [Google Scholar] [CrossRef]
- Ferlay, J.; Steliarova-Foucher, E.; Lortet-Tieulent, J.; Rosso, S.; Coebergh, J.-W.W.; Comber, H.; Forman, D.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur. J. Cancer 2013, 49, 1374–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ljungberg, B.; Bensalah, K.; Canfield, S.; Dabestani, S.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; Lam, T.; Marconi, L.; Merseburger, A.S. EAU guidelines on renal cell carcinoma: 2014 update. Eur. Urol. 2015, 67, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tang, J.; Huang, W.; Wang, F.; Li, P.; Qin, C.; Qin, Z.; Zou, Q.; Wei, J.; Hua, L. The M6A methyltransferase METTL3: Acting as a tumor suppressor in renal cell carcinoma. Oncotarget 2017, 8, 96103–96116. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Antoni, S.; Ferlay, J.; Soerjomataram, I.; Znaor, A.; Jemal, A.; Bray, F. Bladder cancer incidence and mortality: A global overview and recent trends. Eur. Urol. 2017, 71, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Sanli, O.; Dobruch, J.; Knowles, M.A.; Burger, M.; Alemozaffar, M.; Nielsen, M.E.; Lotan, Y. Bladder cancer. Nat. Rev. Dis. Primers 2017, 3, 17022. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Tumor Model | Methodology | Outcome | Sample Size | Author (Ref.) |
|---|---|---|---|---|
| Liver | MeRIP/RIP | METTL3 upregulation associates with poor prognosis | 120 patients, cell lines and animal models | Chen M 2017 [55] |
| m6A-Seq | ||||
| RT-qPCR | ||||
| WB | ||||
| TCGA database | YTHDF1 upregulation associates with poorer stage and survival | 373 patients | Zhao X 2018 [56] | |
| GO and KEGG enrichment analysis * | ||||
| MeRIP/RIP | METTL14 deregulation promotes metastatic spread | 130 patients and animal models | Ma JZ 2017 [57] | |
| RT-qPCR | ||||
| m6A Dot Blot/Immunobloting | ||||
| WB | ||||
| IHC | ||||
| Breast | IHC | FTO overexpression associates with HER-2 positive Breast Cancer | 79 patients | Tan A 2015 [50] |
| WB | Pharmacological inhibition of FTO reduces survival of chemoresistant Inflammatory Breast Cancer cells | Cell lines | Singh B 2016 [51] | |
| IHC | Hypoxia induces cancer stem cell phenotype by ALKBH5-mediated m6A-demethylation | Cell lines | Zhang C 2016 and 2016 [52,53] | |
| RT-qPCR | ||||
| MeRIP/RIP | ||||
| WB | ||||
| Genotyping using custom Illumina array (iCOGS) | SNP in FTP contributes to susceptibility for ER-negative cancer | 6514 patients | Garcia-Closas M 2013 [49] | |
| MeRIP/RIP | Positive feedback loop HBXIP/miR let-7g/METTL3 promotes cancer progression | 24 patients, tissue microarrays (90 breast cancer tissue samples) and cell lines | Cai X 2017 [54] | |
| RT-qPCR | ||||
| m6A Dot Blot/Immunobloting | ||||
| IHC and IF | ||||
| WB | ||||
| Melanoma | GenoMEL * | FTO associates with higher melanoma risk | 1373 patients | Iles MM 2013 [48] |
| Lung | MeRIP/RIP | METTL3 upregulation increases translation of oncogenic pathways | Cell lines | Lin and Choe 2016 [71] |
| m6A-Seq | ||||
| RT-qPCR | ||||
| WB | ||||
| RT-qPCR | METTL3 is targeted by miR-33a attenuating malignant cell proliferation | 32 patients and cell lines | Du M 2016 [72] | |
| WB | ||||
| Brain (Glioblastoma) | MeRIP/RIP | Knockdown of METLL3/METLL14 and FTO inhibition promotes stem cell renewal and tumorigenesis | Cell lines and animal models | Cui Q 2018 [68] |
| m6A-Seq | ||||
| m6A Dot Blot/Immunobloting | ||||
| IF | ||||
| RT-qPCR | ||||
| m6A NorthWestern blot | METTL3 promotes cancer cells maintenance and radioresistance | 57 patients, cell lines and animal models | Visvanathan A 2017 [70] | |
| WB | ||||
| RT-qPCR | ||||
| MeRIP/RIP | ||||
| IHC and IF | ||||
| MeRIP/RIP | ALKBH5 overexpression promotes self-renewal and tumorigenesis through the FOXM1 axis | 604 patients, cell lines and animal models | Zhang S 2017 [69] | |
| m6A-Seq | ||||
| WB | ||||
| IHC and IF | ||||
| RT-qPCR | ||||
| Pancreas | RT-qPCR | METTL3 promotes chemo- and radioresistance | Cell lines | Taketo K 2018 [58] |
| WB | ||||
| RT-qPCR | YTHDF2 is upregulated in cancer and regulates EMT | Cell lines | Chen J, 2017 [59] | |
| IHC | ||||
| WB | ||||
| Biliary tract | cDNA microarray | WTAP promotes migration and invasion | 27 patients, cell lines and animal models | Jo HJ, 2013 [60] |
| RT-qPCR | ||||
| WB | ||||
| IHC | ||||
| Stomach | IHC | FTO overexpression associates with poor prognosis and promotes malignant features | 128 patients and cell lines | Xu D 2017 [64] |
| RT-qPCR | ||||
| WB | ||||
| Cervix | m6A Dot Blot/Immunobloting | Lower m6A levels associate with poor prognosis and malignant features | 286 patients, cell lines and animal models | Wang X 2017 [65] |
| RT-qPCR | ||||
| WB | ||||
| IHC | FTO overexpression leads to chemo- and radioresistance | 30 patients, cell lines and animal models | Zhou S and Bai ZL 2018 [66] | |
| RT-qPCR | ||||
| WB | ||||
| MeRIP/RIP | ||||
| Endometrium | m6A-seq | METTL14 mutation and METTL3 downregulation leads to decreased m6A amount and promotes tumorigenesis by activating AKT signaling | 38 patients, cell lines and animal models | Liu J, 2018 [67] |
| m6A-IP | ||||
| RT-qPCR | ||||
| IHC | ||||
| WB | ||||
| Colorectum | IHC | YTHDF1 overexpression associates with poor prognosis | 63 patients, cell lines and animal models | Nishizawa Y and Kono M 2017 [61] |
| RT-qPCR | ||||
| WB | ||||
| IHC | YTHDC2 overexpression promotes metastases by upregulating HIF-1α | 72 patients and cell lines | Tanabe A 2016 [62] | |
| RT-qPCR | ||||
| WB | ||||
| Leukemia | TCGA database * | Mutations and CNVs in m6A-related genes associate with TP53 mutations and poor prognosis in AML patients | 191 patients | Kwok CT 2017 [73] |
| MeRIP/RIP/ChIP | METTL3 maintains leukemic state | Cell lines and animal models | Barbieri I and Tzelepis K 2017 [75] | |
| ChIP-seq | ||||
| WB | ||||
| RT-qPCR | ||||
| Flow cytometry | ||||
| m6A-seq/RNA-seq | METTL14 promotes leukemogenesis and inhibits hematopoietic stem cell differentiation | Cell lines and animal models | Weng H 2018 [74] | |
| CLIP | ||||
| ChIP | ||||
| WB | ||||
| RT-qPCR | ||||
| Flow cytometry | ||||
| m6A-seq/RNA-seq | FTO promotes leukemogenesis by regulating the ASB2/RARA axis | 100 patients, cell lines and animal models | Li Z 2017 [80] | |
| ChIP | ||||
| WB | ||||
| RT-qPCR | ||||
| m6A Dot Blot/Immunobloting | ||||
| Flow cytometry | ||||
| WB | WTAP promotes leukemic cells proliferation and blocks differentiation | 511 patients, cell lines and animal models | Bansal H 2014 [77] | |
| IP | ||||
| RNA-seq |
| Tumor Model | Sample Size | Most Frequently Deregulated (% of Cases) | Related Alterations (logOR) | Clinicopathological Associations | Survival Impact |
|---|---|---|---|---|---|
| Prostate | 499 tumors | VIRMA (18) | VIRMA + YTHDF3 (co-occurrence, >2) | ↑VIRMA and YTHDF3 in stages III/IV (vs. stage II) | No |
| YTHDF3 (13) | |||||
| (498 patients) | ↑VIRMA and YTHDF3 in GG2-5 (vs. GG1) | ||||
| YTHDC2 (11) | |||||
| Testis | 156 tumors | VIRMA (52) | VIRMA + YTHDF3 (co-occurrence, >3) | ↑VIRMA, YTHDF3, METTL4, ALKBH5 and YTHDC1 in SEs (vs. NSTs) | Yes (METTL4, WTAP, YTHDF1) |
| ↓METTL14 in SEs (vs. NSTs) | |||||
| (150 patients) | YTHDF3 (48) | ↑VIRMA, YTHDF3 and METTL4 in stage I (vs. stages II/III) | |||
| Kidney | 897 tumors | YTHDC2 (21) and RBM15B (14) in ccRCC | VIRMA + YTHDF3 and RBM15B + YTHDC2 (co-occurrence, >3) | ↓VIRMA and YTHDC2 in chRCC and pRCC (vs. ccRCC) | Yes (VIRMA, YTHDC2, RBM15B) |
| VIRMA (17) and HNRNPA2B1 (17) in chRCC | ↑RBM15B in chRCC and pRCC (vs. ccRCC) | ||||
| (895 patients) | |||||
| METTL16 (19), YTHDF1 (19) and RBM15B (14) in pRCC | ↑RBM15B and YTHDC2 in stages II–IV (vs. stage I) | ||||
| Bladder | 413 tumors | VIRMA (29) | METTL14 + YTHDC1 and | ↑VIRMA, METTL4 and YTHDF3 in High Grade tumors (vs. Low Grade tumors) | Yes (WTAP) |
| YTHDF1 (27) | |||||
| ↑VIRMA in non-papillary tumors (vs. papillary tumors) | |||||
| METTL4 (21) | METTL3 + HNRNPC (co-occurrence, 2.3 for all) | ||||
| (412 patients) | YTHDF3 (14) | ||||
| ↑YTHDC1 in papillary tumors (vs. non-papillary tumors) ↑YTHDC1 in stages I/II (vs. stages III/IV) | |||||
| RBM15 (13) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lobo, J.; Barros-Silva, D.; Henrique, R.; Jerónimo, C. The Emerging Role of Epitranscriptomics in Cancer: Focus on Urological Tumors. Genes 2018, 9, 552. https://doi.org/10.3390/genes9110552
Lobo J, Barros-Silva D, Henrique R, Jerónimo C. The Emerging Role of Epitranscriptomics in Cancer: Focus on Urological Tumors. Genes. 2018; 9(11):552. https://doi.org/10.3390/genes9110552
Chicago/Turabian StyleLobo, João, Daniela Barros-Silva, Rui Henrique, and Carmen Jerónimo. 2018. "The Emerging Role of Epitranscriptomics in Cancer: Focus on Urological Tumors" Genes 9, no. 11: 552. https://doi.org/10.3390/genes9110552
APA StyleLobo, J., Barros-Silva, D., Henrique, R., & Jerónimo, C. (2018). The Emerging Role of Epitranscriptomics in Cancer: Focus on Urological Tumors. Genes, 9(11), 552. https://doi.org/10.3390/genes9110552