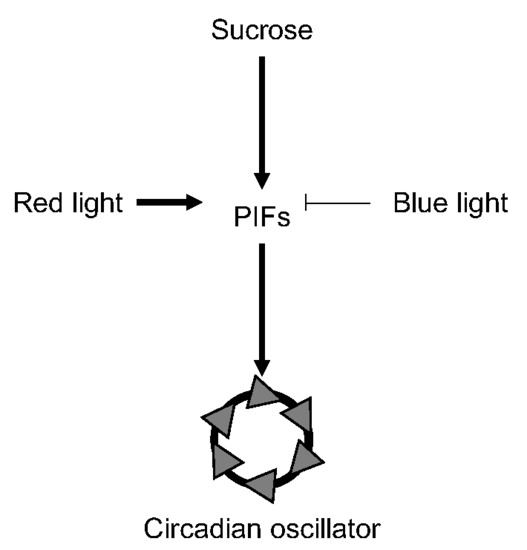
PIF-Mediated Sucrose Regulation of the Circadian Oscillator is Light Quality and Temperature Dependent
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Bioluminescence Assays
2.3. Leaf Movement Assays
2.4. Sucrose Pulses
2.5. RNA Isolation and Analysis
3. Results
3.1. Light-Dependent Roles for PHYTOCHROME INTERACTING FACTOR Regulation of Sucrose Signaling to the Oscillator
3.2. PHYTOCHROME INTERACTING FACTOR-Mediated Sucrose Signaling Is Temperature Dependent
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Alabadi, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Mas, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Harmer, S.L. Wheels within wheels: The plant circadian system. Trends Plant Sci. 2014, 19, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Greenham, K.; McClung, C.R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 2015, 16, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Más, P. Circadian clock function in Arabidopsis thaliana: Time beyond transcription. Trends Cell Biol. 2008, 18, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.A.; Shalit-Kaneh, A.; Chu, D.N.; Hsu, P.Y.; Harmer, S.L. The REVEILLE clock genes inhibit growth of juvenile and adult plants by control of cell size. Plant Physiol. 2017, 173, 2308–2322. [Google Scholar] [CrossRef]
- Nolte, C.; Staiger, D. RNA around the clock—Regulation at the RNA level in biological timing. Front. Plant Sci. 2015, 6, 311. [Google Scholar] [CrossRef]
- Romanowski, A.; Yanovsky, M.J. Circadian rhythms and post-transcriptional regulation in higher plants. Front. Plant Sci. 2015, 6, 437. [Google Scholar] [CrossRef]
- Yakir, E.; Hilman, D.; Hassidim, M.; Green, R.M. CIRCADIAN CLOCK ASSOCIATED1 transcript stability and the entrainment of the circadian clock in Arabidopsis. Plant Physiol. 2007, 145, 925–932. [Google Scholar] [CrossRef]
- Más, P.; Kim, W.Y.; Somers, D.E.; Kay, S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003, 426, 567–570. [Google Scholar] [CrossRef]
- Baudry, A.; Ito, S.; Song, Y.H.; Strait, A.A.; Kiba, T.; Lu, S.; Henriques, R.; Pruneda-Paz, J.L.; Chua, N.H.; Tobin, E.M.; et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell. 2010, 22, 606–622. [Google Scholar] [CrossRef]
- Yakir, E.; Hilman, D.; Harir, Y.; Green, R.M. Regulation of output from the plant circadian clock. FEBS J. 2007, 274, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Thomson, A.J.; McWatters, H.G. SENSITIVE TO FREEZING6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol. 2008, 148, 293–303. [Google Scholar] [CrossRef]
- Haydon, M.J.; Mielczarek, O.; Robertson, F.C.; Hubbard, K.E.; Webb, A.A. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 2013, 502, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Haydon, M.J.; Mielczarek, O.; Frank, A.; Román, Á.; Webb, A.A. Sucrose and ethylene signaling interact to modulate the circadian clock. Plant Physiol. 2017, 175, 947–958. [Google Scholar] [CrossRef]
- Frank, A.; Matiolli, C.C.; Viana, A.J.C.; Hearn, T.H.; Kusakina, J.; Belbin, F.E.; Newman, D.W.; Yochikawa, A.; Cano-Ramirez, D.L.; Chembath, A.; et al. Circadian entrainment in Arabidopsis by the sugar-responsive transcription factor bZIP63. Curr. Biol. 2018, 28, 2597–2606. [Google Scholar] [CrossRef]
- Shor, E.; Paik, I.; Kangisser, S.; Green, R.; Huq, E. Phytochrome Interacting Factors mediate metabolic control of the circadian system in Arabidopsis. New Phytol. 2017, 215, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Paik, I.; Kathare, P.K.; Kim, J.-I.; Huq, E. Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 2017, 10, 1035–1046. [Google Scholar] [CrossRef]
- Kumar, S.V.; Lucyshyn, D.; Jaeger, K.E.; Alós, E.; Alvey, E.; Harberd, N.P.; Wigge, P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012, 484, 242–245. [Google Scholar] [CrossRef] [Green Version]
- Leivar, P.; Monte, E.; Oka, Y.; Liu, T.; Carle, C.; Castillon, A.; Huq, E.; Quail, P.H. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 2008, 18, 1815–1823. [Google Scholar] [CrossRef]
- Zhu, L.; Bu, Q.; Xu, X.; Paik, I.; Huang, X.; Hoecker, U.; Deng, X.W.; Huq, E. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun. 2015, 6, 7245. [Google Scholar] [CrossRef] [Green Version]
- Park, E.; Kim, J.; Lee, Y.; Shin, J.; Oh, E.; Chung, W.I.; Liu, J.R.; Choi, G. Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 2004, 45, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef] [PubMed]
- Plautz, J.D.; Straume, M.; Stanewsky, R.; Jamison, C.F.; Brandes, C.; Dowse, H.B.; Hall, J.C.; Kay, S.A. Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythm. 1997, 12, 204–217. [Google Scholar] [CrossRef] [PubMed]
- Green, R.M.; Tobin, E.M. Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl. Acad. Sci. USA 1999, 96, 4176–4179. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Nozue, K.; Covington, M.F.; Duek, P.D.; Lorrain, S.; Fankhauser, C.; Harmer, S.L.; Maloof, J.N. Rhythmic growth explained by coincidence between internal and external cues. Nature 2007, 448, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Koini, M.A.; Alvey, L.; Allen, T.; Tilley, C.A.; Harberd, N.P.; Whitelam, G.C.; Franklin, K.A. High temperature- mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 2009, 19, 408–413. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Oh, E.; Wang, T.; Wang, Z.Y. TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat Commun. 2016, 7, 13692. [Google Scholar] [CrossRef]
- Spitschan, M.; Aguirre, G.K.; Brainard, D.H.; Sweeney, A.M. Variation of outdoor illumination as a function of solar elevation and light pollution. Sci. Rep. 2016, 6, 26756. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.M.; Zeeman, S.C.; Smith, S.M. Starch degradation. Annu. Rev. Plant Biol. 2005, 56, 73–98. [Google Scholar] [CrossRef]
- Devlin, P.F.; Kay, S.A. Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 2000, 12, 2499–2510. [Google Scholar] [CrossRef] [PubMed]
- Somers, D.E.; Devlin, P.F.; Kay, S.A. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 1998, 282, 1488–1490. [Google Scholar] [CrossRef] [PubMed]
- Pedmale, U.V.; Huang, S.S.; Zander, M.; Cole, B.J.; Hetzel, J.; Ljung, K.; Reis, P.A.; Sridevi, P.; Nito, K.; Nery, J.R.; et al. Cryptochromes Interact Directly with PIFs to Control Plant Growth in Limiting Blue Light. Cell 2016, 164, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Li, X.; Guo, Y.; Chu, J.; Fang, S.; Yan, C.; Noel, J.P.; Liu, H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 2016, 113, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Monte, E. PIFs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.Y.; Fujiwara, S.; Suh, S.S.; Kim, J.; Kim, Y.; Han, L.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 2007, 449, 356–360. [Google Scholar] [CrossRef]
- Kim, J.; Geng, R.; Gallenstein, R.A.; Somers, D.E. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 2013, 140, 4060–4069. [Google Scholar] [CrossRef] [Green Version]
- Stavang, J.A.; Gallego-Bartolome, J.; Gomez, M.D.; Yoshida, S.; Asami, T.; Olsen, J.E.; Garcia-Martinez, J.L.; Alabadi, D.; Blazquez, M.A. Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 2009, 60, 589–601. [Google Scholar] [CrossRef] [Green Version]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef] [Green Version]
- Gould, P.D.; Locke, J.C.; Larue, C.; Southern, M.M.; Davis, S.J.; Hanano, S.; Moyle, R.; Milich, R.; Putterill, J.; Millar, A.J.; et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 2006, 18, 1177–1187. [Google Scholar] [CrossRef] [PubMed]
- Salomé, P.A.; McClung, C.R. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 2005, 17, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Arana, M.V.; Tognacca, R.S.; Estravis-Barcalá, M.; Sánchez, R.A.; Botto, J.F. Physiological and molecular mechanisms underlying the integration of light and temperature cues in Arabidopsis thaliana seeds. Plant Cell Environ. 2017, 40, 3113–3121. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Qi, L.; Li, Y.; Chu, J.; Li, C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012, 8, e1002594. [Google Scholar] [CrossRef] [PubMed]
- Viczian, A.; Kircher, S.; Fejes, E.; Millar, A.J.; Schafer, E.; Kozma-Bognar, L.; Nagy, F. Functional characterization of phytochrome interacting factor 3 for the Arabidopsis thaliana circadian clockwork. Plant Cell Physiol. 2005, 46, 1591–1602. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shor, E.; Potavskaya, R.; Kurtz, A.; Paik, I.; Huq, E.; Green, R. PIF-Mediated Sucrose Regulation of the Circadian Oscillator is Light Quality and Temperature Dependent. Genes 2018, 9, 628. https://doi.org/10.3390/genes9120628
Shor E, Potavskaya R, Kurtz A, Paik I, Huq E, Green R. PIF-Mediated Sucrose Regulation of the Circadian Oscillator is Light Quality and Temperature Dependent. Genes. 2018; 9(12):628. https://doi.org/10.3390/genes9120628
Chicago/Turabian StyleShor, Ekaterina, Raya Potavskaya, Ayelet Kurtz, Inyup Paik, Enamul Huq, and Rachel Green. 2018. "PIF-Mediated Sucrose Regulation of the Circadian Oscillator is Light Quality and Temperature Dependent" Genes 9, no. 12: 628. https://doi.org/10.3390/genes9120628
APA StyleShor, E., Potavskaya, R., Kurtz, A., Paik, I., Huq, E., & Green, R. (2018). PIF-Mediated Sucrose Regulation of the Circadian Oscillator is Light Quality and Temperature Dependent. Genes, 9(12), 628. https://doi.org/10.3390/genes9120628