The Effect of Primary, Secondary, and Tertiary Wastewater Treatment Processes on Antibiotic Resistance Gene (ARG) Concentrations in Solid and Dissolved Wastewater Fractions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
2.2. Collection of Hollow Fiber Filtration (HFF) Concentrated Wastewater Samples
2.3. Separation of Solids and Dissolved Fractions
2.4. DNA Extraction
2.5. AdvIPC:pGEM-T and AdvIPC:pSMART Vectors
2.6. Quantitative PCR Primers/Probes and Plasmids
2.7. Pilot-Scale Dual-Media Filtration
2.8. Statistical Analyses
3. Results
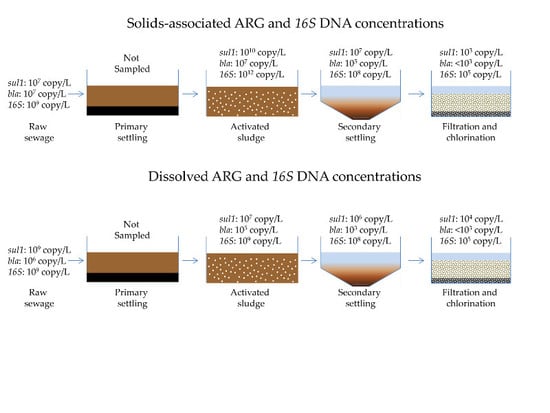
3.1. Quantification of Dissolved and Solids-Associated ARGs and Bacterial 16S DNA Through Primary, Secondary and Tertiary Wastewater Treatment Processes
3.1.1. Antibiotic Resistance Gene Concentrations throughout a Full-Scale WRP
3.1.2. Determination of Total Bacteria Biomass Concentrations in Different Wastewater Treatment Processes by qPCR
3.2. Tertiary Wastewater Treatment Processes did not Result in Positive Selection for blaSHV/TEM or sul1 ARGs
3.3. Evaluation of Tertiary Filtration and Disinfection Processes for the Removal of ARGs
4. Discussion
5. Conclusions
- The full-scale tertiary stage WRP reduced concentrations of sul1 by approximately four-log10 from the raw sewage. In addition, the blaSHV/TEM ARG was reduced to below detectable limits in the final effluent (removal of greater than three log10).
- The percentage of ARGs that partitioned with the solids and dissolved phases differed between treatment processes.
- Positive selection for sul1 or blaSHV/TEM ARGs, in reference to the total bacterial biomass, was not observed throughout the treatment process.
- Tertiary media filtration and chlorine disinfection were the most effective treatment processes with respect to ARG reductions.
- Pilot-scale dual-media filter experiments demonstrated that tertiary filtration enhanced chlorine mediated reduction of an ARG containing plasmid compared to chlorine treatment of secondary effluent.
- This data demonstrated that tertiary filtration and disinfection can result in additional removal of ARGs compared to non-filtered disinfected secondary effluent.
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Mainous, A.G., 3rd; Diaz, V.A.; Matheson, E.M.; Gregorie, S.H.; Hueston, W.J. Trends in hospitalizations with antibiotic-resistant infections: U.S., 1997–2006. Public Health Rep. 2011, 126, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Phelps, C.E. Bug/drug resistance: Sometimes less is more. Med. Care 1989, 27, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, E.A.; Seyfried, E.E.; McMahon, K.D. Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 2007, 41, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Grabow, W.O.; Prozesky, O.W. Drug resistance of coliform bacteria in hospital and city sewage. Antimicrob. Agents Chemother. 1973, 3, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Grabow, W.O.K.; Prozesky, O.W.; Smith, L.S. Drug resistant coliforms call for review of water quality standards. Water Res. 1974, 8, 1–9. [Google Scholar] [CrossRef]
- Linton, K.B.; Richmond, M.H.; Bevan, R.; Gillespie, W.A. Antibiotic resistance and R factors in coliform bacilli isolated from hospital and domestic sewage. J. Med. Microbiol. 1974, 7, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Niemi, M.; Sibakov, M.; Niemela, S. Antibiotic resistance among different species of fecal coliforms isolated from water samples. Appl. Environ. Microbiol. 1983, 45, 79–83. [Google Scholar] [PubMed]
- Reinthaler, F.F.; Posch, J.; Feierl, G.; Wnst, G.; Haas, D.; Ruckenbauer, G.; Mascher, F.; Marth, E. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 2003, 37, 1685–1690. [Google Scholar] [CrossRef]
- Chee-Sanford, J.C.; Aminov, R.I.; Krapac, I.J.; Garrigues-Jeanjean, N.; Mackie, R.I. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 2001, 67, 1494–1502. [Google Scholar] [CrossRef] [PubMed]
- Koike, S.; Krapac, I.G.; Oliver, H.D.; Yannarell, A.C.; Chee-Sanford, J.C.; Aminov, R.I.; Mackie, R.I. Monitoring and source tracking of tetracycline resistance genes in lagoons and groundwater adjacent to swine production facilities over a 3-year period. Appl. Environ. Microbiol. 2007, 73, 4813–4823. [Google Scholar] [CrossRef] [PubMed]
- Sayah, R.S.; Kaneene, J.B.; Johnson, Y.; Miller, R. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 2005, 71, 1394–1404. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, H.; Stoob, K.; Hamscher, G.; Smit, E.; Seinen, W. Tetracyclines and tetracycline resistance in agricultural soils: Microcosm and field studies. Microb. Ecol. 2006, 51, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Martinez, J.L.; Canton, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Feary, T.W.; Sturtevant, A.B.J.; Lankford, J. Antibiotic-resistant coliforms in fresh and salt water. Arch. Environ. Health 1972, 25, 215–220. [Google Scholar] [PubMed]
- Goni-Urriza, M.; Capdepuy, M.; Arpin, C.; Raymond, N.; Caumette, P.; Quentin, C. Impact of an urban effluent on antibiotic resistance of riverine Enterobacteriaceae and Aeromonas spp. Appl. Environ. Microbiol. 2000, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Goñi-Urriza, M.; Pineau, L.; Capdepuy, M.; Roques, C.; Caumette, P.; Quentin, C. Antimicrobial resistance of mesophilic Aeromonas spp. Isolated from two european rivers. J. Antimicrob. Chemother. 2000, 46, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Iwane, T.; Urase, T.; Yamamoto, K. Possible impact of treated wastewater discharge on incidence of antibiotic resistant bacteria in river water. Water Sci. Technol. 2001, 43, 91–99. [Google Scholar] [PubMed]
- Smith, H.W. Incidence of river water of Escherichia coli containing r factors. Nature 1970, 228, 1286–1288. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Aga, D.S.; Jensen, J.N.; Weber, A.S. Effect of sequencing batch reactor operation on presence and concentration of tetracycline-resistant organisms. Water Environ. Res. 2007, 79, 2287–2297. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Marrs, C.F.; Simon, C.; Xi, C. Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci. Total Environ. 2009, 407, 3702–3706. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A.; Arabi, M.; Storteboom, H.N. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ. Sci. Technol. 2012, 46, 11541–11549. [Google Scholar] [CrossRef] [PubMed]
- Martins da Costa, P.; Vaz-Pires, P.; Bernardo, F. Antimicrobial resistance in Enterococcus spp. Isolated in inflow, effluent and sludge from municipal sewage water treatment plants. Water Res. 2006, 40, 1735–1740. [Google Scholar] [CrossRef] [PubMed]
- LaPara, T.M.; Burch, T.R.; McNamara, P.J.; Tan, D.T.; Yan, M.; Eichmiller, J.J. Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into Duluth-Superior Harbor. Environ. Sci. Technol. 2011, 45, 9543–9549. [Google Scholar] [CrossRef] [PubMed]
- Czekalski, N.; Berthold, T.; Caucci, S.; Egli, A.; Burgmann, H. Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into lake geneva, Switzerland. Front. Microbiol. 2012, 3, 106. [Google Scholar] [CrossRef] [PubMed]
- Bockelmann, U.; Dorries, H.H.; Ayuso-Gabella, M.N.; Salgot de Marcay, M.; Tandoi, V.; Levantesi, C.; Masciopinto, C.; Van Houtte, E.; Szewzyk, U.; Wintgens, T.; et al. Quantitative PCR monitoring of antibiotic resistance genes and bacterial pathogens in three European artificial groundwater recharge systems. Appl. Environ. Microbiol. 2009, 75, 154–163. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency. Guidelines for Water Reuse; U.S. Environmental Protection Agency: Washington, DC, USA, 2004; p. 450.
- Rose, J.B.; Farrah, S.R.; Harwood, V.J.; Levine, A.; Lukasik, J.; Menendez, P.; Scott, T.M. Reduction of Pathogens, Indicator Bacteria, and Alternative Indicators by Wastewater Treatment and Reclamation Processes; Water Environment Research Foundation: Alexandria, VA, USA, 2004. [Google Scholar]
- Asano, T. Wastewater Reclamation and Reuse: Water Quality Management Library; Taylor & Francis: Milton Park, UK, 1998. [Google Scholar]
- Mazel, D.; Davies, J. Antibiotic resistance in microbes. Cell Mol. Life Sci. 1999, 56, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yang, X.; Li, J.; Lv, N.; Liu, F.; Wu, J.; Lin, I.Y.; Wu, N.; Weimer, B.C.; Gao, G.F.; et al. The bacterial mobile resistome transfer network connecting the animal and human microbiomes. Appl. Environ. Microbiol. 2016, 82, 6672–6681. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 2009, 11, 2970–2988. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. Biol. Sci. 2009, 276, 2521–2530. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef] [PubMed]
- Bhullar, K.; Waglechner, N.; Pawlowski, A.; Koteva, K.; Banks, E.D.; Johnston, M.D.; Barton, H.A.; Wright, G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 2012, 7, e34953. [Google Scholar] [CrossRef] [PubMed]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Munck, C.; Albertsen, M.; Telke, A.; Ellabaan, M.; Nielsen, P.H.; Sommer, M.O.A. Limited dissemination of the wastewater treatment plant core resistome. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negreanu, Y.; Pasternak, Z.; Jurkevitch, E.; Cytryn, E. Impact of treated wastewater irrigation on antibiotic resistance in agricultural soils. Environ. Sci. Technol. 2012, 46, 4800–4808. [Google Scholar] [CrossRef] [PubMed]
- Pei, R.; Kim, S.C.; Carlson, K.H.; Pruden, A. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 2006, 40, 2427–2435. [Google Scholar] [CrossRef] [PubMed]
- Harms, G.; Layton, A.C.; Dionisi, H.M.; Gregory, I.R.; Garrett, V.M.; Hawkins, S.A.; Robinson, K.G.; Sayler, G.S. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 2003, 37, 343–351. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency. Method A: Enterococci in Water by Taqman Quantitative Polymerase Chain Reaction (qPCR) Assay; EPA-821-R-10-004; U.S. EPA: Washington, DC, USA, 2010; Volume 2010.
- Jothikumar, N.; Cromeans, T.L.; Hill, V.R.; Lu, X.; Sobsey, M.D.; Erdman, D.D. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 2005, 71, 3131–3136. [Google Scholar] [CrossRef] [PubMed]
- Malorny, B.; Paccassoni, E.; Fach, P.; Bunge, C.; Martin, A.; Helmuth, R. Diagnostic real-time PCR for detection of Salmonella in food. Appl. Environ. Microbiol. 2004, 70, 7046–7052. [Google Scholar] [CrossRef] [PubMed]
- Association, A.P.H. Standard Methods for the Examination of Water and Wastewater; General Books: Vancouver, BC, Canada, 2013. [Google Scholar]
- Hicks, L.A.; Bartoces, M.G.; Roberts, R.M.; Suda, K.J.; Hunkler, R.J.; Taylor, T.H., Jr.; Schrag, S.J. US outpatient antibiotic prescribing variation according to geography, patient population, and provider specialty in 2011. Clin. Infect. Dis. 2015, 60, 1308–1316. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M. Beta-lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 1995, 8, 557–584. [Google Scholar] [PubMed]
- Yang, Y.; Zhang, T.; Zhang, X.-X.; Liang, D.-W.; Zhang, M.; Gao, D.-W.; Zhu, H.-G.; Huang, Q.-G.; Fang, H.H. Quantification and characterization of β-lactam resistance genes in 15 sewage treatment plants from East Asia and North America. Appl. Microbiol. Biotechnol. 2012, 95, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.; Boucher, Y.; Labbate, M.; Holmes, A.; Krishnan, S.; Holley, M.; Stokes, H.W. The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 2008, 190, 5095–5100. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, A.W.; Crook, J.; Anderson, M.A.; Bull, R.J.; Drewes, J.E.; Haas, C.N.; Jakubowski, W.; McCarty, P.L.; Nelson, J.B.R.; Sedlak, D.L.; et al. Expert Panel Final Report: Evaluation of the Feasibility of Developing Uniform Water Recycling Criteria for Direct Potable Reuse; California State Water Resources Control Board: Sacramento, CA, USA, 2016; pp. 165–194. [Google Scholar]
- Chen, H.; Zhang, M. Effects of advanced treatment systems on the removal of antibiotic resistance genes in wastewater treatment plants from Hangzhou, China. Environ. Sci. Technol. 2013, 47, 8157–8163. [Google Scholar] [CrossRef] [PubMed]
- Lachmayr, K.L.; Kerkhof, L.J.; Dirienzo, A.G.; Cavanaugh, C.M.; Ford, T.E. Quantifying nonspecific TEM beta-lactamase (blaTEM) genes in a wastewater stream. Appl. Environ. Microbiol. 2009, 75, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.; Wong, K.; Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011, 45, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, M.; Zhang, X.; Fang, H.H. Tetracycline resistance genes and tetracycline resistant lactose-fermenting Enterobacteriaceae in activated sludge of sewage treatment plants. Environ. Sci. Technol. 2009, 43, 3455–3460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhuang, Y.; Geng, J.; Ren, H.; Zhang, Y.; Ding, L.; Xu, K. Inactivation of antibiotic resistance genes in municipal wastewater effluent by chlorination and sequential UV/chlorination disinfection. Sci. Total Environ. 2015, 512–513, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2011, 2, 158. [Google Scholar] [CrossRef] [PubMed]
- Baur, B.; Hanselmann, K.; Schlimme, W.; Jenni, B. Genetic transformation in freshwater: Escherichia coli is able to develop natural competence. Appl. Environ. Microbiol. 1996, 62, 3673–3678. [Google Scholar] [PubMed]
- Guo, M.T.; Yuan, Q.B.; Yang, J. Distinguishing effects of ultraviolet exposure and chlorination on the horizontal transfer of antibiotic resistance genes in municipal wastewater. Environ. Sci. Technol. 2015, 49, 5771–5778. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Wuertz, S. Discrimination of viable and dead fecal Bacteroidales bacteria by quantitative PCR with propidium monoazide. Appl. Environ. Microbiol. 2009, 75, 2940–2944. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Jikumaru, A.; Ueno, T.; Sei, K. Inactivation effect of antibiotic-resistant gene using chlorine disinfection. Water 2017, 9, 547. [Google Scholar] [CrossRef]
- Borjesson, S.; Matussek, A.; Melin, S.; Lofgren, S.; Lindgren, P.E. Methicillin-resistant Staphylococcus aureus (MRSA) in municipal wastewater: An uncharted threat? J. Appl. Microbiol. 2010, 108, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Borjesson, S.; Melin, S.; Matussek, A.; Lindgren, P.E. A seasonal study of the mecA gene and Staphylococcus aureus including methicillin-resistant S. aureus in a municipal wastewater treatment plant. Water Res. 2009, 43, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Garcia, S.; Wade, B.; Bauer, C.; Craig, C.; Nakaoka, K.; Lorowitz, W. The effect of wastewater treatment on antibiotic resistance in Escherichia coli and Enterococcus sp. Water Environ. Res. 2007, 79, 2387–2395. [Google Scholar] [CrossRef] [PubMed]
- Nagulapally, S.R.; Ahmad, A.; Henry, A.; Marchin, G.L.; Zurek, L.; Bhandari, A. Occurrence of ciprofloxacin-, trimethoprim-sulfamethoxazole-, and vancomycin-resistant bacteria in a municipal wastewater treatment plant. Water Environ. Res. 2009, 81, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Rijal, G.K.; Zmuda, J.T.; Gore, R.; Abedin, Z.; Granato, T.; Kollias, L.; Lanyon, R. Antibiotic resistant bacteria in wastewater processed by the metropolitan water reclamation district of greater chicago system. Water Sci. Technol. 2009, 59, 2297–2304. [Google Scholar] [CrossRef] [PubMed]
- Luczkiewicz, A.; Fudala-Ksiazek, S.; Jankowska, K.; Quant, B.; Olanczuk-Neyman, K. Diversity of fecal coliforms and their antimicrobial resistance patterns in wastewater treatment model plant. Water Sci. Technol. 2010, 61, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jensen, J.N.; Aga, D.S.; Weber, A.S. Fate of tetracycline resistant bacteria as a function of activated sludge process organic loading and growth rate. Water Sci. Technol. 2007, 55, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time pcr experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Kralik, P.; Ricchi, M. A basic guide to real time PCR in microbial diagnostics: Definitions, parameters, and everything. Front Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef] [PubMed]



| Target | Primer/Probe | Sequence 1 | Reference |
|---|---|---|---|
| blaSHV/TEM | β-Lac-qPCR-F | 5′-GCCATAACCATGAGYGATAAC-3’ | This study |
| β-Lac-qPCR-R | 5′-TTATCRGCAATAAACCAGCC-3’ | ||
| β-Lac-qPCR Probe | 5’FAM-TCATTCAGCTCCGKTTCCCA-BHQ-1-3’ | ||
| sul1 | sul I-FW | 5′-CGCACCGGAAACATCGCTGCAC-3′ | [42] |
| sul I-RV | 5′-TGAAGTTCCGCCGCAAGGCTCG-3′ | ||
| Bacterial 16S rDNA | Bac 1055f | 5′-ATGGCTGTCGTCAGCT-3′ | [43] |
| Bac 1392r | 5′-ACGGGCGGTGTGTAC-3′ | ||
| 16S Taq1115 | 5′-FAM-CAACGAGCGCAACCC-TAMRA-3’ | ||
| Salmon Sperm DNA | Samn qPCR-F | 5′-GGTTTCCGCAGCTGGG-3’ | [44] |
| Samn qPCR-R | 5′-CCGAGCCGTCCTGGTCTA-3’ | ||
| Samn qPCR-Probe | 5′FAM-AGTCGCAGGCGGCCACCGT -BHQ-1-3’ | ||
| AdvIPC | Adv Hex RT-Forward | 5′-GGAYGCCTCGGAGTACCTGAG-3′ | [45] |
| Adv Hex RT-Reverse | 5′-ACiGTGGGGTTTCTRAACTTGTT-3′ | ||
| Adv IPC Probe | 5′Cy5-CACCGACGGCGAGACCGACTTT-BHQ2-3’ | [41] |
| Gene Target | Raw to AS 1 | Raw to SE 1 | Raw to FE 1 | AS to SE 1 | SE to FE 1 |
|---|---|---|---|---|---|
| blaSHV/TEM | 0.83 3 | 2.84 | ≥3.08 2 | 2.01 | ≥0.24 2 |
| sul1 | 1.46 | 2.74 | 4.79 | 1.28 | 2.05 3 |
| 16S | 0.63 | 1.24 | 3.99 | 0.61 | 2.76 |
| Gene Target | Raw to AS 1 | Raw to SE 1 | Raw to FE 1 | AS to SE 1 | SE to FE 1 |
|---|---|---|---|---|---|
| blaSHV/TEM | −0.14 | 2.11 | ≥3.42 2 | 2.25 | ≥1.31 2 |
| sul1 | −2.38 | 0.40 | 3.98 | 2.78 | 3.58 3 |
| 16S | −2.26 | 1.06 | 4.16 | 3.32 | 3.10 3 |
| ARG:16S 1 | Raw Sewage (n = 3) | Activated Sludge (n = 9) | Secondary Effluent (n = 3) 2 | Final Effluent (n = 6) 3 |
|---|---|---|---|---|
| bla:16S solids | 5.8 × 10−3 ± 7.1 × 10−3 | 2.0 × 10−5 ± 1.4 × 10−5 | 1.7 × 10−4 ± 9.8 × 10−5 | BD |
| sul1:16S solids | 2.0 × 10−2 ± 1.7 × 10−2 | 1.3 × 10−2 ± 8.3 × 10−3 | 6.3 × 10−2 ± 5.3 × 10−2 | 2.9 × 10−2 ± 2.9 × 10−2 |
| bla:16S dissolved | 1.1 × 10−3 ± 5.8 × 10−4 | 3.2 × 10−3 ± 4.2 × 10−3 | 3.3 × 10−5 ± 1.7 × 10−5 | BD |
| sul1:16S dissolved | 2.2 × 10−1 ± 3.8 × 10−1 | 4.8 × 10−2 ± 6.0 × 10−2 | 8.2 × 10−3 ± 6.7 × 10−3 | 2.6 × 10−2 ± 2.9 × 10−2 |
| Sample 1 | Average Log Difference (n = 7) 2 | Rank Sum Test p-Value 3 |
|---|---|---|
| SE vs. chlorinated filtrate | >4.4 ± 0.6 | <0.001 |
| SE vs. chlorinated SE | 2.6 ± 1.4 | <0.001 |
| SE vs. filtrate (no chlorine) | 0.7 ± 0.5 | 0.097 |
| Chlorinated SE vs. chlorinated filtrate | >1.8 ± 1.6 | 0.004 |
| Filtrate vs. chlorinated filtrate | >3.7 ± 0.6 | <0.001 |
| Sample 1 | Average Log Difference (n = 8) 2 | Rank Sum Test p-Value 3 |
|---|---|---|
| SE vs. chlorinated filtrate | >5.2 ± 0.9 | <0.001 |
| SE vs. chlorinated SE | 3.4 ± 1.6 | <0.001 |
| SE vs. filtrate (no chlorine) | 0.9 ± 0.3 | 0.007 |
| Chlorinated SE vs. Chlorinated filtrate | >1.8 ± 1.8 | 0.038 |
| Filtrate vs. chlorinated filtrate | >4.3 ± 0.8 | <0.001 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quach-Cu, J.; Herrera-Lynch, B.; Marciniak, C.; Adams, S.; Simmerman, A.; Reinke, R.A. The Effect of Primary, Secondary, and Tertiary Wastewater Treatment Processes on Antibiotic Resistance Gene (ARG) Concentrations in Solid and Dissolved Wastewater Fractions. Water 2018, 10, 37. https://doi.org/10.3390/w10010037
Quach-Cu J, Herrera-Lynch B, Marciniak C, Adams S, Simmerman A, Reinke RA. The Effect of Primary, Secondary, and Tertiary Wastewater Treatment Processes on Antibiotic Resistance Gene (ARG) Concentrations in Solid and Dissolved Wastewater Fractions. Water. 2018; 10(1):37. https://doi.org/10.3390/w10010037
Chicago/Turabian StyleQuach-Cu, Jennipher, Bellanira Herrera-Lynch, Christine Marciniak, Scott Adams, April Simmerman, and Ryan A. Reinke. 2018. "The Effect of Primary, Secondary, and Tertiary Wastewater Treatment Processes on Antibiotic Resistance Gene (ARG) Concentrations in Solid and Dissolved Wastewater Fractions" Water 10, no. 1: 37. https://doi.org/10.3390/w10010037
APA StyleQuach-Cu, J., Herrera-Lynch, B., Marciniak, C., Adams, S., Simmerman, A., & Reinke, R. A. (2018). The Effect of Primary, Secondary, and Tertiary Wastewater Treatment Processes on Antibiotic Resistance Gene (ARG) Concentrations in Solid and Dissolved Wastewater Fractions. Water, 10(1), 37. https://doi.org/10.3390/w10010037