Nitrate Removal and Dynamics of Microbial Community of A Hydrogen-Based Membrane Biofilm Reactor at Diverse Nitrate Loadings and Distances from Hydrogen Supply End
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reactor Configuration
2.2. Synthetic Influent
2.3. Experimental Operation
2.4. Analytical Methods of Aquatic Samples
2.5. Biofilm Sampling and Analysis
3. Results and Discussion
3.1. Denitrification Performance
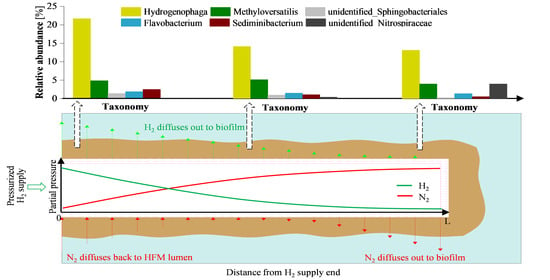
3.2. Microbial Community Analysis
3.3. Predictive Functional Genes
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wu, J.; Yin, Y.; Wang, J. Hydrogen-based membrane biofilm reactors for nitrate removal from water and wastewater. Int. J. Hydrogen Energ. 2018, 43, 1–15. [Google Scholar] [CrossRef]
- Terada, A.; Hibiya, K.; Nagai, J.; Tsuneda, S.; Hirata, A. Nitrogen removal characteristics and biofilm analysis of a membrane-aerated biofilm reactor applicable to high-strength nitrogenous wastewater treatment. J. Biosci. Bioeng. 2003, 95, 170–178. [Google Scholar] [CrossRef]
- Chen, M.; Price, R.M.; Yamashita, Y.; Jaffé, R. Comparative study of dissolved organic matter from groundwater and surface water in the Florida coastal Everglades using multi-dimensional spectrofluorometry combined with multivariate statistics. Appl. Geochem. 2010, 25, 872–880. [Google Scholar] [CrossRef]
- Shen, Y.; Chapelle, F.H.; Strom, E.W.; Benner, R. Origins and bioavailability of dissolved organic matter in groundwater. Biogeochemistry 2015, 122, 61–78. [Google Scholar] [CrossRef]
- Martin, K.J.; Nerenberg, R. The membrane biofilm reactor (MBfR) for water and wastewater treatment: Principles, applications, and recent developments. Bioresour. Technol. 2012, 122, 83–94. [Google Scholar] [CrossRef]
- Terada, A.; Kaku, S.; Matsumoto, S.; Tsuneda, S. Rapid autohydrogenotrophic denitrification by a membrane biofilm reactor equipped with a fibrous support around a gas-permeable membrane. Biochem. Eng. J. 2006, 31, 84–91. [Google Scholar] [CrossRef]
- Lee, K.C.; Rittmann, B.E. A novel hollow-fibre membrane biofilm reactor for autohydrogenotrophic denitrification of drinking water. Water Sci. Technol. 2000, 41, 219–226. [Google Scholar] [CrossRef]
- Tang, Y.; Zhou, C.; Van Ginkel, S.W.; Ontiveros-Valencia, A.; Shin, J.; Rittmann, B.E. Hydrogen permeability of the hollow fibers used in H2-based membrane biofilm reactors. J. Membr. Sci. 2012, 407–408, 176–183. [Google Scholar] [CrossRef]
- Xia, S.; Wang, C.; Xu, X.; Tang, Y.; Wang, Z.; Gu, Z.; Zhou, Y. Bioreduction of nitrate in a hydrogen-based membrane biofilm reactor using CO2 for pH control and as carbon source. Chem. Eng. J. 2015, 276, 59–64. [Google Scholar] [CrossRef]
- Healy, R.W.; Bartos, T.T.; Rice, C.A.; McKinley, M.P.; Smith, B.D. Groundwater chemistry near an impoundment for produced water, Powder River Basin, Wyoming, USA. J. Hydrol. 2011, 403, 37–48. [Google Scholar] [CrossRef]
- Zhu, G.F.; Li, Z.Z.; Su, Y.H.; Ma, J.Z.; Zhang, Y.Y. Hydrogeochemical and isotope evidence of groundwater evolution and recharge in Minqin Basin, Northwest China. J. Hydrol. 2007, 333, 239–251. [Google Scholar] [CrossRef]
- Perez-Calleja, P.; Aybar, M.; Picioreanu, C.; Esteban-Garcia, A.L.; Martin, K.J.; Nerenberg, R. Periodic venting of MABR lumen allows high removal rates and high gas-transfer efficiencies. Water Res. 2017, 121, 349–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, S.; Zhong, F.; Zhang, Y.; Li, H.; Yang, X. Bio-reduction of nitrate from groundwater using a hydrogen-based membrane biofilm reactor. J. Environ. Sci. 2010, 22, 257–262. [Google Scholar] [CrossRef]
- Lee, K.-C.; Rittmann, B.E. Applying a novel autohydrogenotrophic hollow-fiber membrane biofilm reactor for denitrification of drinking water. Water Res. 2002, 36, 2040–2052. [Google Scholar] [CrossRef]
- Zhao, H.-P.; Ontiveros-Valencia, A.; Tang, Y.; Kim, B.-O.; VanGinkel, S.; Friese, D.; Overstreet, R.; Smith, J.; Evans, P.; Krajmalnik-Brown, R.; et al. Removal of multiple electron acceptors by pilot-scale, two-stage membrane biofilm reactors. Water Res. 2014, 54, 115–122. [Google Scholar] [CrossRef]
- Ahmed, T.; Semmens, M.J. The use of independently sealed microporous hollow fiber membranes for oxygenation of water: Model development. J. Membr. Sci. 1992, 69, 11–20. [Google Scholar] [CrossRef]
- Li, H.; Zhou, L.; Lin, H.; Xu, X.; Jia, R.; Xia, S. Dynamic response of biofilm microbial ecology to para-chloronitrobenzene biodegradation in a hydrogen-based, denitrifying and sulfate-reducing membrane biofilm reactor. Sci. Total Environ. 2018, 643, 842–849. [Google Scholar] [CrossRef]
- Long, M.; Ilhan, Z.E.; Xia, S.; Zhou, C.; Rittmann, B.E. Complete dechlorination and mineralization of pentachlorophenol (PCP) in a hydrogen-based membrane biofilm reactor (MBfR). Water Res. 2018, 144, 134–144. [Google Scholar] [CrossRef]
- Li, H.; Zhou, L.; Lin, H.; Zhang, W.; Xia, S. Nitrate effects on perchlorate reduction in a H2/CO2-based biofilm. Sci. Total Environ. 2019, 694, 133564. [Google Scholar] [CrossRef]
- Xia, S.; Liang, J.; Xu, X.; Shen, S. Simultaneous removal of selected oxidized contaminants in groundwater using a continuously stirred hydrogen-based membrane biofilm reactor. J. Environ. Eng. 2013, 25, 96–104. [Google Scholar] [CrossRef]
- Su, X.; Wang, H.; Zhang, Y. Health risk assessment of nitrate contamination in groundwater: A case study of an agricultural area in Northeast China. Water Resour. Manag. 2013, 27, 3025–3034. [Google Scholar] [CrossRef]
- Gu, B.; Ying, G.; Chang, S.X.; Luo, W.; Jie, C. Nitrate in groundwater of China: Sources and driving forces. Glob. Environ. Chang. 2013, 23, 1112–1121. [Google Scholar] [CrossRef]
- Gao, S.; Li, C.; Jia, C.; Zhang, H.; Guan, Q.; Wu, X.; Wang, J.; Lv, M. Health risk assessment of groundwater nitrate contamination: A case study of a typical karst hydrogeological unit in East China. Environ. Sci. Pollut. R. 2020, 27, 9274–9287. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Xu, X.; Xia, S. Effects of sulfate on simultaneous nitrate and selenate removal in a hydrogen-based membrane biofilm reactor for groundwater treatment: Performance and biofilm microbial ecology. Chemosphere 2018, 211, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Xu, X.; Zhou, C.; Wang, C.; Zhou, L.; Rittmann, B.E. Direct delivery of CO2 into a hydrogen-based membrane biofilm reactor and model development. Chem. Eng. J. 2016, 290, 154–160. [Google Scholar] [CrossRef]
- Ontiveros-Valencia, A.; Ziv-El, M.; Zhao, H.-P.; Feng, L.; Rittmann, B.E.; Krajmalnik-Brown, R. Interactions between nitrate-reducing and sulfate-reducing bacteria coexisting in a hydrogen-fed biofilm. Environ. Sci. Technol. 2012, 46, 11289–11298. [Google Scholar] [CrossRef]
- Jiang, M.; Zheng, J.; Perez-Calleja, P.; Picioreanu, C.; Lin, H.; Zhang, X.; Zhang, Y.; Li, H.; Nerenberg, R. New insight into CO2-mediated denitrification process in H2-based membrane biofilm reactor: An experimental and modeling study. Water Res. 2020, 184, 116177. [Google Scholar] [CrossRef]
- Lai, C.-Y.; Yang, X.; Tang, Y.; Rittmann, B.E.; Zhao, H.-P. Nitrate Shaped the Selenate-Reducing Microbial Community in a Hydrogen-Based Biofilm Reactor. Environ. Sci. Technol. 2014, 48, 3395–3402. [Google Scholar] [CrossRef]
- Ding, X.W.; Wei, D.; Guo, W.S.; Wang, B.; Meng, Z.J.; Feng, R.; Du, B.; Wei, Q. Biological denitrification in an anoxic sequencing batch biofilm reactor: Performance evaluation, nitrous oxide emission and microbial community. Bioresour. Technol. 2019, 285, 9. [Google Scholar] [CrossRef]
- Lai, C.-Y.; Wen, L.-L.; Zhang, Y.; Luo, S.-S.; Wang, Q.-Y.; Luo, Y.-H.; Chen, R.; Yang, X.; Rittmann, B.E.; Zhao, H.-P. Autotrophic antimonate bio-reduction using hydrogen as the electron donor. Water Res. 2016, 88, 467–474. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Long, M.; Zeng, C.; Wang, Z.; Xia, S.; Zhou, C. Complete dechlorination and mineralization of para-chlorophenol (4-CP) in a hydrogen-based membrane biofilm reactor (MBfR). J. Clean. Prod. 2020, 276, 123257. [Google Scholar] [CrossRef]
- Lv, P.L.; Shi, L.D.; Dong, Q.Y.; Rittmann, B.; Zhao, H.P. How nitrate affects perchlorate reduction in a methane-based biofilm batch reactor. Water Res. 2020, 171, 115397. [Google Scholar] [CrossRef]
- Pang, Y.; Wang, J. Insight into the mechanism of chemoautotrophic denitrification using pyrite (FeS2) as electron donor. Bioresour. Technol. 2020, 318, 124105. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Tang, Y.; Zhao, H.; Marcus, A.K.; Krajmalnik-Brown, R.; Rittmann, B.E. A steady-state biofilm model for simultaneous reduction of nitrate and perchlorate, part 1: Model development and numerical solution. Environ. Sci. Technol. 2012, 46, 1598–1607. [Google Scholar] [CrossRef]
- Park, H.I.; Choi, Y.-J.; Pak, D. Autohydrogenotrophic Denitrifying microbial community in a glass beads biofilm reactor. Biotechnol. Lett. 2005, 27, 949–953. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhong, F.; Xia, S.; Wang, X.; Li, J. Autohydrogenotrophic denitrification of drinking water using a polyvinyl chloride hollow fiber membrane biofilm reactor. J. Hazard. Mater. 2009, 170, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.-P.; Van Ginkel, S.; Tang, Y.; Kang, D.-W.; Rittmann, B.; Krajmalnik-Brown, R. Interactions between Perchlorate and Nitrate Reductions in the Biofilm of a Hydrogen-Based Membrane Biofilm Reactor. Environ. Sci. Technol. 2011, 45, 10155–10162. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-H.; Choi, O.; Lee, T.-H.; Kim, H.; Sang, B.-I. Pyrosequencing analysis of microbial communities in hollow fiber-membrane biofilm reactors system for treating high-strength nitrogen wastewater. Chemosphere 2016, 163, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Ontiveros-Valencia, A.; Tang, Y.; Krajmalnik-Brown, R.; Rittmann, B.E. Perchlorate reduction from a highly contaminated groundwater in the presence of sulfate-reducing bacteria in a hydrogen-fed biofilm. Biotechnol. Bioeng. 2013, 110, 3139–3147. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Wu, C.; Yang, X.; Zhou, Y.; Zhou, L.; Ran, Y.; Rittmann, B.E. Bioreduction of nitrate in high-sulfate water using a hydrogen-based membrane biofilm reactor equipped with a separate carbon dioxide module. Chem. Eng. J. 2020, 385, 123831. [Google Scholar] [CrossRef]
- Zhao, H.P.; Ontiveros-Valencia, A.; Tang, Y.N.; Kim, B.O.; Ilhan, Z.E.; Krajmalnik-Brown, R.; Rittrnann, B. Using a two-stage hydrogen-based membrane biofilm reactor (MBfR) to achieve complete perchlorate reduction in the presence of nitrate and sulfate. Environ. Sci. Technol. 2013, 47, 1565–1572. [Google Scholar] [CrossRef]
- Ontiveros-Valencia, A.; Ilhan, Z.E.; Kang, D.-W.; Rittmann, B.; Krajmalnik-Brown, R. Phylogenetic analysis of nitrate- and sulfate-reducing bacteria in a hydrogen-fed biofilm. FEMS Microbiol. Ecol. 2013, 85, 158–167. [Google Scholar] [CrossRef]
- Wei, Z.S.; Wang, J.B.; Huang, Z.S.; Xiao, X.L.; Tang, M.R.; Li, B.L.; Zhang, X. Removal of nitric oxide from biomass combustion by thermophilic nitrification-aerobic denitrification combined with catalysis in membrane biofilm reactor. Biomass Bioenerg. 2019, 126, 34–40. [Google Scholar] [CrossRef]
- Quan, Z.-X.; Im, W.-T.; Lee, S.-T. Azonexus caeni sp. nov., a denitrifying bacterium isolated from sludge of a wastewater treatment plant. Int. J. Syst. Evol. Micr. 2006, 56, 1043–1046. [Google Scholar] [CrossRef]
- Qu, J.-H.; Yuan, H.-L. Sediminibacterium salmoneum gen. nov., sp. nov., a member of the phylum Bacteroidetes isolated from sediment of a eutrophic reservoir. Int. J. Syst. Evol. Micr. 2008, 58, 2191–2194. [Google Scholar] [CrossRef] [Green Version]
- Ontiveros-Valencia, A.; Tang, Y.; Krajmalnik-Brown, R.; Rittmann, B.E. Managing the interactions between sulfate-and perchlorate-reducing bacteria when using hydrogen-fed biofilms to treat a groundwater with a high perchlorate concentration. Water Res. 2014, 55, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Kojima, H.; Fukui, M. Complete genomes of freshwater sulfur oxidizers Sulfuricella denitrificans skB26 and Sulfuritalea hydrogenivorans sk43H: Genetic insights into the sulfur oxidation pathway of betaproteobacteria. Syst. Appl. Microbiol. 2014, 37, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Fetzner, S. Biodegradation of Xenobiotics. In Biotechnology; Encyclopedia of Life Support Systems (EOLSS); EOLSS Publishers Co., Ltd.: Oxford, UK, 2002; pp. 215–246. [Google Scholar]
- Rice, A.J.; Park, A.; Pinkett, H.W. Diversity in ABC transporters: Type I, II and III importers. Crit. Rev. Biochem. Mol. 2014, 49, 426–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabba, F.; Picioreanu, C.; Boltz, J.P.; Nerenberg, R. Predicting N2O emissions from nitrifying and denitrifying biofilms: A modeling study. Water Sci. Technol. 2017, 75, 530–538. [Google Scholar] [CrossRef] [PubMed]







| Reductase | KO_Hierarchy | KEGG_Description | N1 | N2 | N3 | D | M | U |
|---|---|---|---|---|---|---|---|---|
| Nitrate reductase | K00370 | nitrate reductase alpha subunit | 12,718 | 18,940 | 16,095 | 15,010 | 14,465 | 14,245 |
| K00371 | nitrate reductase beta subunit | 12,783 | 18,984 | 16,111 | 15,421 | 14,485 | 14,296 | |
| K00374 | nitrate reductase gamma subunit | 8040 | 18,639 | 15,925 | 12,799 | 14,172 | 13,816 | |
| K02567 | nitrate reductase (cytochrome) | 5236 | 501 | 437 | 694 | 372 | 408 | |
| K02568 | nitrate reductase (cytochrome), electron transfer subunit | 5215 | 490 | 423 | 668 | 362 | 399 | |
| Nitrite reductase | K00368 | nitrite reductase (NO-forming) | 3676 | 1837 | 3077 | 7378 | 6180 | 6000 |
| Nitric oxide reductase | K04561 | nitric oxide reductase subunit B | 14,325 | 19,475 | 16,173 | 13,828 | 14,392 | 14,122 |
| Nitrous oxide reductase | K00376 | nitrous-oxide reductase | 10,580 | 1231 | 890 | 1666 | 1152 | 1040 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, M.; Zhang, Y.; Yuan, Y.; Chen, Y.; Lin, H.; Zheng, J.; Li, H.; Zhang, X. Nitrate Removal and Dynamics of Microbial Community of A Hydrogen-Based Membrane Biofilm Reactor at Diverse Nitrate Loadings and Distances from Hydrogen Supply End. Water 2020, 12, 3196. https://doi.org/10.3390/w12113196
Jiang M, Zhang Y, Yuan Y, Chen Y, Lin H, Zheng J, Li H, Zhang X. Nitrate Removal and Dynamics of Microbial Community of A Hydrogen-Based Membrane Biofilm Reactor at Diverse Nitrate Loadings and Distances from Hydrogen Supply End. Water. 2020; 12(11):3196. https://doi.org/10.3390/w12113196
Chicago/Turabian StyleJiang, Minmin, Yuanyuan Zhang, Yuhang Yuan, Yuchao Chen, Hua Lin, Junjian Zheng, Haixiang Li, and Xuehong Zhang. 2020. "Nitrate Removal and Dynamics of Microbial Community of A Hydrogen-Based Membrane Biofilm Reactor at Diverse Nitrate Loadings and Distances from Hydrogen Supply End" Water 12, no. 11: 3196. https://doi.org/10.3390/w12113196
APA StyleJiang, M., Zhang, Y., Yuan, Y., Chen, Y., Lin, H., Zheng, J., Li, H., & Zhang, X. (2020). Nitrate Removal and Dynamics of Microbial Community of A Hydrogen-Based Membrane Biofilm Reactor at Diverse Nitrate Loadings and Distances from Hydrogen Supply End. Water, 12(11), 3196. https://doi.org/10.3390/w12113196