Effects of Hydraulic Retention Time and Influent Nitrate-N Concentration on Nitrogen Removal and the Microbial Community of an Aerobic Denitrification Reactor Treating Recirculating Marine Aquaculture System Effluent
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Aerobic Denitrification Reactor
2.2. Experimental Design
2.3. Water Samples and Quality Analysis
2.4. DNA Sample and DNA Extraction
2.5. Processing of Sequencing Data
2.6. Statistical Analysis
2.7. Statistical Analyses
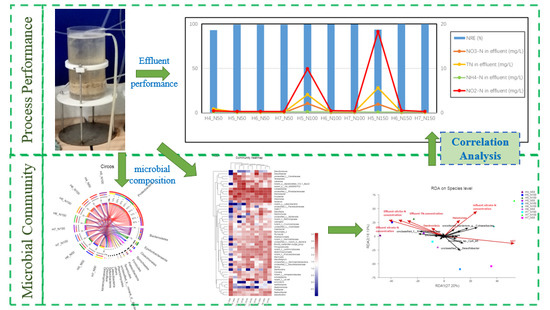
3. Results
3.1. Performance of Nitrogen Removal
3.2. Microbial Community Analysis
3.2.1. Richness and Alpha Diversity Analyses of Bacterial Communities
3.2.2. Bacterial Community Composition at the Phylum and Genus Levels
3.2.3. Community Composition and Response to Environmental Variables
4. Discussion
4.1. Process Performance
4.2. Microbial Community Structure
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Vollenweider, R.A. Scientific Fundamentals of The Eutrophication of Lakes and Flowing Waters, with Particular Reference to Nitrogen and Phosphorus as Factors in Eutrophication; OECD: Paris, France, 1970. [Google Scholar]
- Ryther, J.H.; Dunstan, W.M. Nitrogen, phosphorus, and eutrophication in the coastal marine environment. Science 1971, 171, 1008–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heisler, J.; Gilbert, P.M.; Burkholder, J.M.; Anderson, D.M.; Cochlan, W.; Dennison, W.C.; Dortch, Q.; Gobler, C.J.; Heil, C.A.; Humphries, E.; et al. Eutrophication and harmful algal blooms: A scientific consensus. Harmful Algae 2008, 8, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gobler, C.J.; Burkholder, J.M.; Davis, T.W.; Harke, M.J.; Johengen, T.; Stow, C.A.; Van de Waal, D.B. The dual role of nitrogen supply in controlling the growth and toxicity of cyanobacterial blooms. Harmful Algae 2016, 54, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Boyd, C.E. Water Quality Management for Pond Fish Culture; Elsevier Scientific Publishing Co.: Amsterdam, The Netherlands, 1982. [Google Scholar]
- Camargo, J.A.; Alonso, A.; Salamanca, A. Nitrate toxicity to aquatic animals: A review with new data for freshwater invertebrates. Chemosphere 2005, 58, 1255–1267. [Google Scholar] [CrossRef]
- Davidson, J.; Good, C.; Welsh, C.; Summerfelt, S.T. Comparing the effects of high vs. low nitrate on the health, performance, and welfare of juvenile rainbow trout Oncorhynchus mykiss within water recirculating aquaculture systems. Aquacult. Eng. 2014, 59, 30–40. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Hu, F.; Guo, W.; Hallerman, E.; Song, X.; Huang, Z. Acute and chronic toxicity of nitrate to greenlings (Hexagrammos otakii). J. World Aquac. Soc. in press. [CrossRef]
- Yang, X.; Song, X.; Peng, L.; Hallerman, E.; Huang, Z. Effects of nitrate on aquaculture production, blood and histological markers and liver transcriptome of Oplegnathus punctatus (Temminck et Schlegel, 1846). Aquaculture 2019, 501, 387–396. [Google Scholar] [CrossRef]
- Lekang, O.-I. Ammonia removal. In Aquaculture Engineering; Blackwell Publishing: Oxford, UK, 2007; pp. 121–132. [Google Scholar]
- Van Rijn, J.; Tal, Y.; Schreier, H.J. Denitrification in recirculating systems: Theory and applications. Aquac. Eng. 2006, 34, 364–376. [Google Scholar] [CrossRef]
- Luo, G.; Xu, G.; Gao, J.; Tan, H. Effect of dissolved oxygen on nitrate removal using polycaprolactone as an organic carbon source and biofilm carrier in fixed-film denitrifying reactors. J. Environ. Sci. 2016, 43, 147–152. [Google Scholar] [CrossRef]
- Ding, A.; Zhao, D.; Ding, F.; Du, S.; Lu, H.; Zhang, M.; Zheng, P. Effect of inocula on performance of bio-cathode denitrification and its microbial mechanism. Chem. Eng. J. 2018, 343, 399–407. [Google Scholar] [CrossRef]
- Xu, Z.; Dai, X.; Chai, X. Effect of influent pH on biological denitrification using biodegradable PHBV/PLA blends as electron donor. Biochem. Eng. J. 2018, 131, 24–30. [Google Scholar] [CrossRef]
- Liu, Y.; Li, G.; Chen, Z.; Megharaj, M.; Naidu, R. Removal of nitrate using Paracoccus sp. Yf1 immobilized on bamboo carbon. J. Hazard. Mater. 2012, 229, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Wang, X.; Zang, M.; Zhou, J.; Wang, J.; Guo, W. Degradation pathways and kinetics of anthraquinone compounds along with nitrate removal by a newly isolated Rhodococcus pyridinivorans Gf3 under aerobic conditions. Bioresour. Technol. 2019, 285, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Shi, J.; Ma, F. Aerobic denitrification and biomineralization by a novel heterotrophic bacterium, Acinetobacter sp. H36. Mar. Pollut. Bull. 2017, 116, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Wang, T.; Ye, J.; Zhao, J.; Yang, L.; Wu, P.; Duan, J.L.; Ye, G. Effects of carbon sources and operation modes on the performances of aerobic denitrification process and its microbial community shifts. J. Environ. Manag. 2019, 239, 299–305. [Google Scholar] [CrossRef]
- Ji, B.; Yang, K.; Zhu, L.; Jiang, Y.; Wang, H.; Zhou, J.; Zhang, H. Aerobic denitrification: A review of important advances of the last 30 years. Biotechnol. Bioprocess Eng. 2015, 20, 643–651. [Google Scholar] [CrossRef]
- Hu, Z.; Zhang, J.; Xie, H.; Li, S.; Wang, J.; Zhang, T. Effect of anoxic/aerobic phase fraction on N2O emission in a sequencing batch reactor under low temperature. Bioresour. Technol. 2011, 102, 5486–5491. [Google Scholar] [CrossRef]
- Vacková, L.; Srb, M.; Stloukal, R.; Wanner, J. Comparison of denitrification at low temperature using encapsulated Paracoccus denitrificans, Pseudomonas fluorescens and mixed culture. Bioresour. Technol. 2011, 102, 4661–4666. [Google Scholar] [CrossRef]
- Lee, K.C.; Rittmann, B.E. Effects of pH and precipitation on autohydrogenotrophic denitrification using the hollow-fiber membrane-biofilm reactor. Water Res. 2003, 37, 1551–1556. [Google Scholar] [CrossRef]
- Grommen, R.; Verhaege, M.; Verstraete, W. Removal of nitrate in aquaria by means of electrochemically generated hydrogen gas as electron donor for biological denitrification. Aquac. Eng. 2006, 34, 33–39. [Google Scholar] [CrossRef]
- Lee, N.M.; Welander, T. The effect of different carbon sources on respiratory denitrification in biological wastewater treatment. J. Ferment. Bioeng. 1996, 82, 277–285. [Google Scholar] [CrossRef]
- Yang, W.; He, S.; Han, M.; Wang, B.; Niu, Q.; Xu, Y.; Chen, Y.; Wang, H. Nitrogen removal performance and microbial community structure in the start-up and substrate inhibition stages of an anammox reactor. J. Biosci. Bioeng. 2018, 126, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chu, L. Biological nitrate removal from water and wastewater by solid-phase denitrification process. Biotechnol. Adv. 2016, 34, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zuo, J.; Wang, Y.; Zhao, J.; Tang, L.; Li, Z. Tertiary nitrogen removal for municipal wastewater using a solid-phase denitrifying biofilter with polycaprolactone as the carbon source and filtration medium. Water Res. 2016, 93, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Song, L.; Dai, X.; Chai, X. PHBV polymer supported denitrification system efficiently treated high nitrate concentration wastewater: Denitrification performance, microbial community structure evolution and key denitrifying bacteria. Chemosphere 2018, 197, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Qiu, T.; Xu, Y.; Gao, M.; Han, M.; Wang, X. Bacterial community dynamics in a biodenitrification reactor packed with polylactic acid/poly(3-hydroxybutyrate-co-3-hydroxyvalerate) blend as the carbon source and biofilm carrier. J. Biosci. Bioeng. 2017, 123, 606–612. [Google Scholar] [CrossRef]
- Boopathy, R.; Bonvillain, C.; Fontenot, Q.; Kilgen, M. Biological treatment of low-salinity shrimp aquaculture wastewater using sequencing batch reactor. Int. Biodeter. Biodegrad. 2007, 59, 16–19. [Google Scholar] [CrossRef]
- Fontenot, Q.; Bonvillain, C.; Kilgen, M.; Boopathy, R. Effects of temperature, salinity, and carbon: Nitrogen ratio on sequencing batch reactor treating shrimp aquaculture wastewater. Bioresour. Technol. 2007, 98, 1700–1703. [Google Scholar] [CrossRef]
- Borges, M.T.; Sousa, A.; De Marco, P.; Matos, A.; Hönigová, P.; Castro, P.M. Aerobic and anoxic growth and nitrate removal capacity of a marine denitrifying bacterium isolated from a recirculation aquaculture system. Microb. Ecol. 2008, 55, 107–118. [Google Scholar] [CrossRef]
- Gutierrez-Wing, M.T.; Malone, R.F.; Rusch, K.A. Evaluation of polyhydroxybutyrate as a carbon source for recirculating aquaculture water denitrification. Aquac. Eng. 2012, 51, 36–43. [Google Scholar] [CrossRef]
- Zhu, S.M.; Deng, Y.L.; Ruan, Y.J.; Guo, X.S.; Shi, M.M.; Shen, J.Z. Biological denitrification using poly (butylene succinate) as carbon source and biofilm carrier for recirculating aquaculture system effluent treatment. Bioresour. Technol. 2015, 192, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Estrada, A.M.; Gollas-Galvan, T.; Martinez-Cordova, L.R.; Martinez-Porchas, M. Predictive functional profiles using metagenomics 16S rRNA data: A novel functional approach to understanding the microbial ecology or aquaculture systems. Rev. Aquacult. 2019, 11, 234–245. [Google Scholar] [CrossRef] [Green Version]
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Nat. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaud, L.; Lo Giudice, A.; Troussellier, M.; Smedile, F.; Bruni, V.; Blancheton, J.P. Phylogenetic characterization of the heterotrophic bacterial communities inhabiting a marine recirculating aquaculture system. J. Appl. Microbiol. 2009, 107, 1935–1946. [Google Scholar] [CrossRef] [PubMed]
- Keuter, S.; Kruse, M.; Lipski, A.; Spieck, E. Relevance of Nitrospira for nitrite oxidation in a marine recirculation aquaculture system and physiological features of a Nitrospira marina-like isolate. Environ. Microbiol. 2011, 13, 2536–2547. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wan, R.; Song, X.; Liu, Y.; Hallerman, E.; Dong, D.; Zhai, J.M.; Zhang, H.S.; Sun, L.Y. Metagenomic analysis shows diverse, distinct bacterial communities in biofilters among different marine recirculating aquaculture systems. Aquac. Int. 2016, 24, 1393–1408. [Google Scholar] [CrossRef]
- Rud, I.; Kolarevic, J.; Holan, A.B.; Berget, I.; Calabrese, S.; Terjesen, B.F. Deep-sequencing of the bacterial microbiota in commercial-scale recirculating and semi-closed aquaculture systems for Atlantic salmon post-smolt production. Aquac. Eng. 2017, 78, 50–62. [Google Scholar] [CrossRef]
- Brailo, M.; Schreier, H.J.; McDonald, R.; Maršić-Lučić, J.; Gavrilović, A.; Pećarević, M.; Jug-Dujaković, J. Bacterial community analysis of marine recirculating aquaculture system bioreactors for complete nitrogen removal established from a commercial inoculum. Aquaculture 2019, 503, 198–206. [Google Scholar] [CrossRef]
- Ahn, Y.H. Sustainable nitrogen elimination biotechnologies: A review. Process Biochem. 2006, 41, 1709–1721. [Google Scholar] [CrossRef]
- Zhang, R.C.; Xu, X.J.; Chen, C.; Shao, B.; Zhou, X.; Yuan, Y.; Lee, D.J.; Ren, N.Q. Bioreactor performance and microbial community analysis of autotrophic denitrification under micro-aerobic condition. Sci. Total Environ. 2019, 647, 914–922. [Google Scholar] [CrossRef]
- Jiang, Y.L.; Huang, Z.T.; Song, X.F. Carbon metabolism characteristics of microbial community in aerobic denitrification reactor. Ocean. Limnol. 2018, 49, 331–339. (In Chinese) [Google Scholar]
- Huang, Z.T.; Song, X.; Jiang, Y.; Hallerman, E. Characterization of a novel haloduric heterotrophic nitrifying and aerobically denitrifying bacterium, Halomonas sp. Z8 as a potential bioagent for wastewater treatment. J. Environ. Biol. 2020, 41, 43–52. [Google Scholar] [CrossRef]
- APHA (American Public Health Association), American Water Works Association, Water Environment Federation. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 2005; Volume 21, pp. 258–259. [Google Scholar]
- Peng, X.; Zhang, S.; Li, L.; Zhao, X.; Ma, Y.; Shi, D. Long-term high-solids anaerobic digestion of food waste: Effects of ammonia on process performance and microbial community. Bioresour. Technol. 2018, 262, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ji, X.; Wang, J.; Yang, D.; Shen, Y.; Chen, C.; Qian, F.Y.; Wu, P. Microbial community response to influent shift and lowering temperature in a two-stage mainstream deammonification process. Bioresour. Technol. 2018, 262, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Meth. 2013, 10, 996. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2014, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.; Zhou, Z.; Pang, H.; Zheng, Y.; Chen, L.; Jiang, L.M.; Zhao, X. Correlation of microbial community structure with pollutants removal, sludge reduction and sludge characteristics in micro-aerobic side-stream reactor coupled membrane bioreactors under different hydraulic retention times. Bioresour. Technol. 2018, 260, 177–185. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, M.O.; Gauch, H.G. Detrended correspondence analysis: An improved ordination technique. In Classification and Ordination; Springer: Dordrecht, The Netherlands, 1980; pp. 47–58. [Google Scholar]
- Van Den Wollenberg, A.L. Redundancy analysis an alternative for canonical correlation analysis. Psychometrika 1977, 42, 207–219. [Google Scholar] [CrossRef]
- Fruchterman, T.M.J.; Reingold, E.M. Graph drawing by force-directed placement. Softw. Pract. Exper. 1991, 21, 1129–1164. [Google Scholar] [CrossRef]
- Colt, J.; Lamoureux, J.; Patterson, R.; Rogers, G. Reporting standards for biofilter performance studies. Aquac. Eng. 2006, 34, 377–388. [Google Scholar] [CrossRef]
- Kim, B.-S.; Lim, Y.W.; Chun, J. Sphingopyxis marina sp. nov. and Sphingopyxis litoris sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 2008, 58, 2415–2419. [Google Scholar] [CrossRef] [Green Version]
- Luján-Facundo, M.J.; Fernández-Navarro, J.; Alonso-Molina, J.L.; Amorós- Muñoz, I.; Moreno, Y.; Mendoza-Roca, J.A.; Pastor-Alcañiz, L. The role of salinity on the changes of the biomass characteristics and on the performance of an OMBR treating tannery wastewater. Water Res. 2018, 142, 129–137. [Google Scholar] [CrossRef]
- Simon, M.; Scheuner, C.; Meier-Kolthoff, J.P.; Brinkhoff, T.; Wagner-Döbler, I.; Ulbrich, M.; Klenk, H.P.; Schomburg, D.; Petersen, J.; Göker, M. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 2017, 11, 1483–1499. [Google Scholar] [CrossRef]
- Selje, N.; Simon, M.; Brinkhoff, T. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 2004, 427, 445–448. [Google Scholar] [CrossRef]
- Giebel, H.A.; Kalhoefer, D.; Lemke, A.; Thole, S.; Gahl-Janssen, R.; Simon, M.; Brinkhoff, T. Distribution of Roseobacter RCA and SAR11 lineages in the North Sea and characteristics of an abundant RCA isolate. ISME J. 2011, 5, 8. [Google Scholar] [CrossRef]
- Wemheuer, B.; Wemheuer, F.; Hollensteiner, J.; Meyer, F.; Voget, S.; Daniel, R. The green impact: Bacterioplankton response towards a phytoplankton spring bloom in the southern North Sea assessed by comparative metagenomic and metatranscriptomic approaches. Front. Microbiol. 2015, 6, 805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fadhlaoui, K.; Ben, H.W.; Postec, A.; Fauque, G.; Hamdi, M.; Ollivier, B.; Fardeau, M.L. Fusibacter fontis sp. nov., a sulfur-reducing, anaerobic bacterium isolated from a mesothermic Tunisian spring. Int. J. Syst. Evol. Microbiol. 2015, 65, 3501. [Google Scholar] [CrossRef] [PubMed]
- Priyanka, P.; Arun, A.B.; Rekha, P.D. Sulfated exopolysaccharide produced by Labrenzia sp. PRIM-30, characterization and prospective applications. Int. J. Biol. Macromol. 2014, 69, 290–295. [Google Scholar]
- Okamoto, T.; Maruyama, A.; Imura, S.; Takeyama, H.; Naganuma, T. Comparative phylogenetic analyses of Halomonas variabilis and related organisms based on 16S rRNA, gyrB and ectBC gene sequences. Syst. Appl. Microbiol 2004, 27, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Quillaguamán, J.; Hashim, S.; Bento, F.; Mattiasson, B.; Hatti-Kaul, R. Poly(ß-hydroxybutyrate) production by a moderate halophile, Halomonas boliviensis LC1 using starch hydrolysate as substrate. J. Appl. Microbiol. 2005, 99, 151. [Google Scholar] [CrossRef]
- Zinatizadeh, A.A.L.; Ghaytooli, E. Simultaneous nitrogen and carbon removal from wastewater at different operating conditions in a moving bed biofilm reactor (MBBR): Process modeling and optimization. J. Taiwan Inst. Chem. Eng. 2015, 53, 98–111. [Google Scholar] [CrossRef]
- Wang, X.M.; Wang, J.L. Nitrate removal from groundwater using solid-phase denitrification process without inoculating with external microorganisms. Int. J. Environ. Sci. Technol. 2013, 10, 955–960. [Google Scholar] [CrossRef] [Green Version]
- Ovez, B.; Ozgen, S.; Yuksel, M. Biological denitrification in drinking water using Glycyrrhiza glabra and Arunda donax as the carbon source. Process Biochem. 2006, 41, 1539–1544. [Google Scholar] [CrossRef]
- Zhu, W.J.; Yang, Y.; Qiao, Y.M.; Ran, T. Effect of hydraulic retention time on purification efficiency of natural aerated biological filter bed. Ecol. Sci. 2013, 32, 224–229. [Google Scholar]
- Qiu, L.P.; Ma, J.; Zhang, L.X. Effects of hydraulic residence time on treatment efficiency and operation characteristics of biological aerated filter. Environ. Pollut. Prevent. Cont. 2004, 26, 433–436. [Google Scholar]
- Körner, H.; Zumft, W.G. Expression of denitrification enzymes in response to the dissolved oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl. Environ. Microbiol. 1989, 55, 1670–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, L.; Wang, J. Denitrification performance and biofilm characteristics using biodegradable polymers PCL as carriers and carbon source. Chemosphere 2013, 91, 1310–1316. [Google Scholar] [CrossRef] [PubMed]
- Hamlin, H.J.; Michaels, J.T.; Beaulaton, C.M.; Graham, W.F.; Dutt, W.; Steinbach, P.; Losordo, T.M.; Schrader, K.K.; Main, K.L. Comparing denitrification rates and carbon sources in commercial scale upflow denitrification biological filters in aquaculture. Aquac. Eng. 2008, 38, 79–92. [Google Scholar] [CrossRef]
- Kiss, H.; Lang, E.; Lapidus, A.; Kiss, H.; Lang, E.; Lapidus, A.; Copeland, A.; Nolan, M.; Del Rio, T.G.; Han, C. Complete genome sequence of Denitrovibrio acetiphilus type strain (N2460 T). Stand. Genom. Sci. 2010, 2, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torno, J.; Naas, C.; Schroeder, J.P.; Schulz, C. Impact of hydraulic retention time, back flushing intervals, and C/N ratio on the SID-reactor denitrification performance in marine RAS. Aquaculture 2018, 496, 112–122. [Google Scholar] [CrossRef]
- Yin, H.; Yang, C.; Jia, Y.; Chen, H.; Gu, X. Dual removal of phosphate and ammonium from high concentrations of aquaculture wastewaters using an efficient two-stage infiltration system. Sci. Total Environ. 2018, 635, 936–946. [Google Scholar] [CrossRef]
- Obaja, D.; Mace, S.; Mata-Alvarez, J. Biological nutrient removal by a sequencing batch reactor (SBR) using an internal organic carbon source in digested piggery wastewater. Bioresour. Technol. 2005, 96, 7–14. [Google Scholar] [CrossRef]
- Elefsiniotis, P.; Wareham, D.G.; Smith, M.O. Use of volatile fatty acids from an acid phase digester for denitrification. J. Biotechnol. 2004, 114, 289–297. [Google Scholar] [CrossRef]
- Xu, Z.; Dai, X.; Chai, X. Effect of different carbon sources on denitrification performance, microbial community structure and denitrification genes. Sci. Total Environ. 2018, 634, 195–204. [Google Scholar] [CrossRef]
- Nicolella, C.; van Loosdrecht, M.C.M.; Heijnen, J.J. Wastewater treatment with particulate biofilm reactors. J. Biotechnol. 2000, 80, 1–33. [Google Scholar] [CrossRef]
- Cunningham, A.B.; Characklis, W.G.; Aberdeen, F.; Crawford, D. Influence of biofilm accumulation on porous media hydrodynamics. Environ. Sci. Technol. 1991, 25, 1305–1311. [Google Scholar] [CrossRef]
- Kondaveeti, S.; Lee, S.H.; Park, H.D.; Min, B. Bacterial communities in a bioelectrochemical denitrification system: The effects of supplemental electron acceptors. Water Res. 2014, 51, 25–36. [Google Scholar] [CrossRef]
- Lu, H.J.; Chandran, K.; Stensel, D. Microbial ecology of denitrification in biological wastewater treatment. Water Res. 2014, 64, 237–254. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, Z.; Yang, Y.; Mei, X.; Wu, Z. Correlating microbial community structure and composition with aeration intensity in submerged membrane bioreactors by high-throughput pyrosequencing. Water Res. 2013, 47, 859–869. [Google Scholar] [CrossRef]
- Nakasaki, K.; Tran, L.T.H.; Idemoto, Y.; Abe, M.; Rollon, A.P. Comparison of organic matter degradation and microbial community during thermophilic composting of two different types of anaerobic sludge. Bioresour. Technol. 2009, 100, 676–682. [Google Scholar] [CrossRef]
- Lam, P.; Kuypers, M.M. Microbial nitrogen cycling processes in oxygen minimum zones. Annu. Rev. Mar. Sci. 2011, 15, 317–345. [Google Scholar] [CrossRef]
- Li, Y.; Tang, K.; Zhang, L.; Zhao, Z.; Xie, X.; Chen, C.T.; Wang, D.; Jiao, N.; Zhang, Y. Coupled carbon, sulfur, and nitrogen cycles mediated by microorganisms in the water column of a shallow-water hydrothermal ecosystem. Front. Microbiol. 2018, 13, 2718. [Google Scholar] [CrossRef] [Green Version]
- Baek, K.; Choi, A.; Kang, I.; Cho, J.C. Celeribacter marinus sp. nov., isolated from coastal seawater. Int. J. Syst. Evol. Microbiol. 2014, 64, 1323–1327. [Google Scholar] [CrossRef]
- Arahal, D.R.; Lenkunberru, I.; Gonzalez, J.M.; Pascual, J.; Pujalte, M.J.; Pedros-Alio, C.; Pinhassi, J. Neptuniibacter caesariensis gen. nov., sp. nov., a novel marine genome-sequenced gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 2007, 57, 1000–1006. [Google Scholar] [CrossRef] [Green Version]
- Vaidya, B.; Kumar, R.; Sharma, G.; Srinivas, T.N.; Kumar, P.A. Xanthomarina gelatinilytica gen. nov., sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 2015, 65, 3926–3932. [Google Scholar] [CrossRef]
- Huang, X.-F.; Liu, Y.J.; Dong, J.-D.; Qu, L.-Y.; Zhang, Y.-Y.; Wang, F.-Z.; Tian, X.-P.; Zhang, S. Mangrovibacterium diazotrophicum gen. nov., sp. nov., a nitrogen-fixing bacterium isolated from a mangrove sediment, and proposal of Prolixibacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Ben Hania, W.; Joseph, M.; Bunk, B.; Sproer, C.; Klenk, H.P.; Fardeau, M.L.; Spring, S. Characterization of the first cultured representative of a Bacteroidetes clade specialized on the scavenging of cyanobacteria. Environ. Microbiol. 2017, 19, 1134–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letelier-Gordo, C.O.; Herreros, M.M. Denitrifying granules in a marine upflow anoxic sludge bed (UASB) reactor. Aquac. Eng. 2019, 84, 42–49. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J.L.; Nguyen, H.; Meyer, R.L.; Nielsen, P.H. Identification of glucose fermenting bacteria in a full-scale enhanced biological phosphorus removal plant by stable isotope probing. Microbiology 2012, 158, 1818–1825. [Google Scholar] [CrossRef] [Green Version]
- McGenity, T.; Van Der Meer, J.R.; de Lorenzo, V. Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin, Germany, 2010. [Google Scholar]
- Fernandez, N.; Sierra-Alvarez, R.; Field, J.A.; Amils, R.; Sanz, J.L. Microbial community dynamics in a chemolithotrophic denitrification reactor inoculated with methanogenic granular sludge. Chemosphere 2008, 70, 462–474. [Google Scholar] [CrossRef]
- Dannenberg, S.; Kroder, M.; Dilling, W.; Cypionka, H. Oxidation of H2, organic compounds and inorganic sulfur compounds coupled to reduction of O2 or nitrate by sulfate-reducing bacteria. Arch. Microbiol. 1992, 158, 93–99. [Google Scholar] [CrossRef]
- Shen, Z.; Zhou, Y.; Liu, J.; Xiao, Y.; Cao, R.; Wu, F. Enhanced removal of nitrate using starch/PCL blends as solid carbon source in a constructed wetland. Bioresour. Technol. 2015, 175, 239–244. [Google Scholar] [CrossRef]
- Du, R.; Cao, S.B.; Li, B.K.; Niu, M.; Wang, S.Y.; Peng, Y.Z. Performance and microbial community analysis of a novel DEAMOX based on partial-denitrification and anammox treating ammonia-N and nitrate wastewaters. Water Res. 2017, 108, 46–56. [Google Scholar] [CrossRef]
- Wu, W.; Yang, L.; Wang, J. Denitrification performance and microbial diversity in a packed-bed bioreactor using PCL as carbon source and biofilm carrier. Appl. Microbiol. Biotechnol. 2013, 97, 2725–2733. [Google Scholar] [CrossRef]




| Experimental Phase 1 | Times (d) | Q (mL/min) | Influent Nitrate-N Concentration (mg/L) | HRT (h) |
|---|---|---|---|---|
| H4_N50 | 1–40 | 9.16 | 50 | 4 |
| H5_N50 | 41–64 | 7.33 | 50 | 5 |
| H6_N50 | 65–88 | 6.11 | 50 | 6 |
| H7_N50 | 89–112 | 5.23 | 50 | 7 |
| H5_N100 | 113–136 | 7.33 | 100 | 5 |
| H6_N100 | 137–160 | 6.11 | 100 | 6 |
| H7_N100 | 161–184 | 5.23 | 100 | 7 |
| H5_N150 | 185–208 | 7.33 | 150 | 5 |
| H6_N150 | 209–232 | 6.11 | 150 | 6 |
| H7_N150 | 233–255 | 5.23 | 150 | 7 |
| Sample | Nitrate-N in Influent (mg/L) | Ammonia-N in Effluent (mg/L) | Nitrate-N in Effluent (mg/L) | Nitrite-N in Effluent (mg/L) | Temperature | Dissolved Oxygen | pH | Salinity | TN in Effluent (mg/L) | NRE (%) | NRR (g·N·m−3·d−1) | No. of Samples |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H4_N50 | 50.23 ± 1.74 | 0.18 ± 0.42 | 4.04 ± 0.49a | 0.43 ± 0.53a | 25.56 ± 0.28 | 6.39 ± 0.16 | 7.87 ± 0.05 | 34.21 ± 0.03 | 4.48 ± 1.01a | 92.90% | 697.39 ± 27.75a | 20 |
| H5_N50 | 52.69 ± 2.21 | 0.16 ± 0.25 | 0.31 ± 0.24d | 0.31 ± 0.14a | 25.60 ± 0.29 | 6.38 ± 0.15 | 7.88 ± 0.07 | 34.18 ± 0.05 | 0.62 ± 0.21d | 99.38% | 625.09 ± 25.19b | 12 |
| H6_N50 | 52.14 ± 1.57 | 0.18 ± 0.26 | 0.29 ± 0.24d | 0.31 ± 0.14a | 25.56 ± 0.29 | 6.39 ± 0.17 | 7.87 ± 0.06 | 34.21 ± 0.04 | 0.61 ± 0.21d | 99.42% | 511.96 ± 19.72c | 12 |
| H7_N50 | 52.66 ± 1.58 | 0.21 ± 0.09 | 0.31 ± 0.24d | 0.31 ± 0.14a | 25.60 ± 0.33 | 6.48 ± 0.16 | 7.88 ± 0.08 | 34.18 ± 0.06 | 0.63 ± 0.21d | 99.38% | 442.51 ± 16.55d | 12 |
| H5_N100 | 102.32 ± 3.61 | 0.23 ± 0.08 | 10.63 ± 0.91b | 9.91 ± 1.01d | 25.56 ± 0.30 | 6.39 ± 0.18 | 7.87 ± 0.07 | 34.31 ± 0.15 | 20.54 ± 1.42e | 99.51% | 1094.98 ± 51.25e | 12 |
| H6_N100 | 100.83 ± 4.23 | 0.22 ± 0.11 | 0.45 ± 0.31d | 0.47 ± 0.31a | 25.58 ± 0.31 | 6.48 ± 0.17 | 7.86 ± 0.09 | 34.38 ± 0.27 | 0.92 ± 0.46d | 99.55% | 973.49 ± 41.68f | 12 |
| H7_N100 | 100.61 ± 3.99 | 0.21 ± 0.10 | 0.44 ± 0.34d | 0.41 ± 0.34a | 25.56 ± 0.31 | 6.39 ± 0.19 | 7.87 ± 0.08 | 34.21 ± 0.06 | 0.85 ± 0.53d | 99.56% | 855.07 ± 31.92g | 12 |
| H5_N150 | 152.85 ± 4.71 | 0.18 ± 0.41 | 9.75 ± 3.37b | 18.3 ± 2.51e | 25.60 ± 0.28 | 6.48 ± 0.18 | 7.88 ± 0.10 | 34.18 ± 0.08 | 28.09 ± 4.93f | 93.50% | 1718.56 ± 54.91h | 12 |
| H6_N150 | 150.43 ± 2.39 | 0.19 ± 0.12 | 0.39 ± 0.32d | 0.51 ± 0.24a | 25.56 ± 0.32 | 6.39 ± 0.20 | 7.88 ± 0.10 | 34.21 ± 0.07 | 0.91 ± 0.35d | 99.74% | 1512.22 ± 44.99i | 12 |
| H7_N150 | 151.35 ± 3.28 | 0.18 ± 0.39 | 0.37 ± 0.29d | 0.29 ± 0.26a | 25.61 ± 0.28 | 6.48 ± 0.19 | 7.87 ± 0.09 | 34.18 ± 0.09 | 0.66 ± 0.26d | 99.75% | 1285.52 ± 29.35j | 12 |
| Sample | Reads | Coverage | Richness | Diversity | Evenness | ||||
|---|---|---|---|---|---|---|---|---|---|
| Sobs | ACE | Chao | Shannon | Simpson | Shannoneven | Simpsoneven | |||
| H4_N50 | 21,333 | 0.998 | 174 | 198.5 | 214.6 | 3.62 | 0.047 | 0.739 | 0.140 |
| H5_N50 | 21,568 | 0.998 | 215 | 247.7 | 247.2 | 3.77 | 0.042 | 0.692 | 0.072 |
| H6_N50 | 31,914 | 0.997 | 259 | 302.5 | 299.5 | 3.92 | 0.036 | 0.705 | 0.079 |
| H7_N50 | 22,379 | 0.998 | 211 | 241.4 | 242.5 | 3.27 | 0.075 | 0.607 | 0.046 |
| H5_N100 | 25,098 | 0.998 | 217 | 255.6 | 248.5 | 3.61 | 0.055 | 0.660 | 0.053 |
| H6_N100 | 27,859 | 0.997 | 197 | 239.8 | 233.1 | 3.53 | 0.061 | 0.666 | 0.053 |
| H7_N100 | 24,728 | 0.998 | 227 | 253.1 | 245.0 | 3.74 | 0.041 | 0.694 | 0.079 |
| H5_N150 | 22,255 | 0.998 | 207 | 243.4 | 242.6 | 3.60 | 0.055 | 0.669 | 0.057 |
| H6_N150 | 26,958 | 0.998 | 156 | 191.1 | 184.0 | 3.26 | 0.069 | 0.633 | 0.062 |
| H7_N150 | 24,170 | 0.998 | 174 | 200.3 | 203.3 | 3.19 | 0.085 | 0.608 | 0.041 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Yang, X.; Hallerman, E.; Jiang, Y.; Huang, Z. Effects of Hydraulic Retention Time and Influent Nitrate-N Concentration on Nitrogen Removal and the Microbial Community of an Aerobic Denitrification Reactor Treating Recirculating Marine Aquaculture System Effluent. Water 2020, 12, 650. https://doi.org/10.3390/w12030650
Song X, Yang X, Hallerman E, Jiang Y, Huang Z. Effects of Hydraulic Retention Time and Influent Nitrate-N Concentration on Nitrogen Removal and the Microbial Community of an Aerobic Denitrification Reactor Treating Recirculating Marine Aquaculture System Effluent. Water. 2020; 12(3):650. https://doi.org/10.3390/w12030650
Chicago/Turabian StyleSong, Xiefa, Xiaohan Yang, Eric Hallerman, Yuli Jiang, and Zhitao Huang. 2020. "Effects of Hydraulic Retention Time and Influent Nitrate-N Concentration on Nitrogen Removal and the Microbial Community of an Aerobic Denitrification Reactor Treating Recirculating Marine Aquaculture System Effluent" Water 12, no. 3: 650. https://doi.org/10.3390/w12030650
APA StyleSong, X., Yang, X., Hallerman, E., Jiang, Y., & Huang, Z. (2020). Effects of Hydraulic Retention Time and Influent Nitrate-N Concentration on Nitrogen Removal and the Microbial Community of an Aerobic Denitrification Reactor Treating Recirculating Marine Aquaculture System Effluent. Water, 12(3), 650. https://doi.org/10.3390/w12030650